1 Department of Biotechnology, Maharishi Markandeshwar (Deemed to be University), Mullana, India
2Department of Biotechnology, Mukand Lal National College, Yamuna Nagar, India.
3Institute of Plant Sciences, Agricultural Research Organisations, Rishon LeZion, Israel
4Department of Applied Science & Humanities, Seth Jai Parkash Mukand Lal Institute
of Engineering and Technology, Radaur, India.
5Department of Biochemistry, International Institute of Veterinary Education and Research,
Lala Lajpat Rai University of Veterinary and Animal Sciences, Rohtak, India.
Corresponding author email: hardeep.biotech@gmail.com
Article Publishing History
Received: 19/07/2021
Accepted After Revision: 17/09/2021
Diabetes is considered to be a fatal disease as it brings many malfunctioning to the body. Currently used therapy is found to possess lots of side effects. Hence, we need to turn to more potent and safer options. An alternative choice can be phytochemicals of common use that will be cost-effective and safe. Therefor the aim of this research was to identify and analyze plant based metabolites as glusocidase inhibitors. This enzyme is known to enhance carbohydrate digestion and responsible to increase the level of glucose in blood circulation. Hence inhibitors of glucosidase are in limelight all over the world to regulate type-2 diabetes by reducing carbohydrate digestion and absorption. In the present study two phytochemicals prangenidin and columbin were selected as ligands. α-glucosidase was chosen as ligand’s receptor from Protein Data Bank. Molecular docking of ligand & receptor was carried out by using PyRx molecular docking software.
The docking results are found to be quite promising with the binding affinity of -6.6 kcal to -7.7 kcal and -6.4 to -8.1 kcal for prangenidin and columbin respectively. It can be concluded from the results of the present study that these phytochemicals have a high affinity to bind with α-glucosidase and may be used in the near future to deal with diabetes type II. In addition, the present study may be helpful to initiate in vitro/ in vivo research by using these phytochemicals. Such evidence based researches will provide a platform to start clinical trials for the treatment of fatal diseases like diabetes.
Binding Affinity, Diabetes Type II, Molecular Docking, Phytochemicals, Α-Glucosidase
Tuli H. S, Sood S, Upadhyay S. K, Kumar P, Seth P, Vashishth A. In silico Molecular Docking of Α-Glucosidase with Prangenidin and Columbin as an Anti-Hyperglycemic Strategy. Biosc.Biotech.Res.Comm. 2021;14(3).
Tuli H.S, Sood S, Upadhyay S.K, Kumar P, Seth P, Vashishth A. In silico Molecular Docking of Α-Glucosidase with Prangenidin and Columbin as an Anti-Hyperglycemic Strategy. Biosc.Biotech.Res.Comm. 2021;14(3). Available from: <a href=”https://bit.ly/3iSxhgi“>https://bit.ly/3iSxhgi</a>
Copyright © Tuli et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
α-Glucosidase (EC 3.2.1.20) is considered as a key enzyme that regulates the conversion of carbohydrates into glucose. Therefore, it is known to regulate the blood sugar level and control type–II diabetes (Venable and Aschenbrenner 2002; Park et al. 2008; Gamblin et al. 2009; Rawling et al. 2009).
Previous studies suggested that inhibition of α-glucosidase activity may be utilized to design promising therapy for type-II diabetes. Scientists have synthesized several molecules as an inhibitor of α-glucosidase. However, synthetic molecules are found to possess many side effects such as abdominal discomfort, diarrhea, flatulence, and hepatotoxicity (Campbell et al. 2000; Krentz and Bailey, 2005; Hsiao et al. 2006; Nathan et al. 2006; Rehman et al. 2019; Kato-Schwartz et al. 2020).
As a result, molecular docking tools may provide a promising route to design and identify novel inhibitor for the α-glucosidase enzyme (Raghu et al. 2019; O’Keefe et al. 2019). There have been several α-glucosidase inhibitors including acarbose, voglibose, and miglitol obtained from natural sources with clinical implications (Lefebvre and Scheen 1994; Scott and Spencer 2000; Playford et al. 2013). However, very few α-glucosidase inhibitors are commercially available.
Therefore the search of novel natural inhibitors of the α-glucosidase enzyme is still going on. In recent years, projects undertaken to discover potent non-sugar based α-glucosidase inhibitors from natural sources have received tremendous attention (Chang et al. 2013; Mata et al. 2013). A majority of the compounds reported contain flavonoid and terpene ring structures (Humphries et al. 1986; Hwangseo et al. 2008; Rudd et al. 2008; Yin et al. 2014; Yousuf et al. 2020; Kato-Schwartz et al. 2020).
The ligand of the present study i.e., Prangenidin is also known as Alloimperatorin and belongs to the coumarin compound. It is an active chemical content of the plant Aegl emarmelos (Indian name- Bael) and known to be extracted from Angelica dahurica. It is found to have anti-inflammatory, anti-oxidant, and anti-neoplastic activity (Chen et al. 2008; Li et al. 2016).
Another ligand of the present study is Columbin, an organic heterocyclic compound. It is an active chemical content of the plant Tinospora cordifolila (Indian name- guduchi) and known to have anti-inflammatory and anti-cancer properties (Abdelwahab et al. 2012). Furthermore, a search on the pub-med timeline has shown an increasing trend of research on diabetes (Chung et al. 2019; Zhang et al. 2020; Chen et al. 2021). Therefore, in the present study, Prangenidin and Columbin ligands were docked with the α-glucosidase enzyme to find out their binding potential with enzyme and to explore future outcomes to control diabetes type –II.
MATERIAL AND METHODS
We performed molecular docking between ligands (Prangenidin and Columbin) & enzyme α-glucosidase. The enzyme molecule was obtained from PDB in its PDB format and its PDB ID is 5DKY. Protein structure was viewed in BIOVIA Discovery Studio Visualizer, and hetero-atoms, water molecules, ligand groups and nucleic acid groups were removed. Polar hydrogens were added and nonpolar hydrogens were merged. Missing atoms were checked and repaired before applying Kollman charges. The macromolecule was saved in PDBQT file format for the further application.
The ligand structures were obtained from chemsketch in its SAG format and were checked for drug likeness and its physiochemical properties by using Lipinski rule of 5. The software used for molecular docking was PyRx, a useful virtual screening software for computational drug discovery to screen libraries of compounds against potential drug targets. PyRx includes docking wizards with an easy-to-use set up which makes it a valuable tool for Computer-Aided Drug Design (Tuli et al. 2021).
RESULTS AND DISCUSSION
Molecular Docking or QSAR is a technique to explore the binding affinity of the ligand to its receptor. In this method, a scoring function used to find the highest affinity position for the tested ligand in the active site of the receptor. In molecular docking, it has been known that the lesser the energy relates to high affinities between ligand and receptor. QSAR analysis of molecules has gained importance to find a convenient ligand before performing expensive and time-consuming wet laboratory experiments.
The physio-chemical properties of the selected ligands (Prangenidin & Columbin) have described in table 1. Prangenidin on docking with α-glucosidase gave the best binding energy affinity of -7.7. It interacted with two amino acids of α-glucosidase, to be exact at GLY-228 and GLU-271. Table 2 represented α-glucosidase amino acids interactions with ligand Prangenidin and the distance between the interacted ligand poles with an amino acid. Whereas Table 3 represented the complete molecular docking results of ligand and receptor α-glucosidase. The docked pose of ligand and α-glucosidase has been shown in Figure 1.
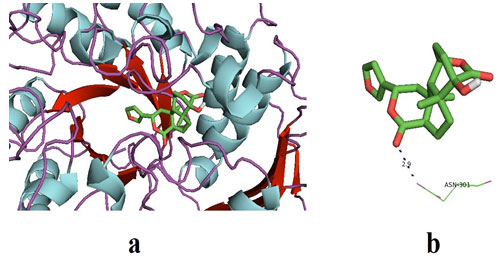
Columbin on docking with α-glucosidase gave the best binding energy affinity of -8.1. It interacted with two amino acids of α-glucosidase, to be exact at ASN-301. The amino acids of α-glucosidase interacted with columbin, and complete docking results were shown in table 4. Furthermore, the interaction between the active poles of the ligand and the amino acid of the receptor (α-glucosidase) i.e ASN-301 was observed with a distance of 2.9Å.
It represents strong H-bonds formation between ligand and receptor. Figure 2 represented the docked pose of Columbin and α-glucosidase receptors in the software. The results of the present study are found to be consistent with previously published docking results that showed binding energies in the range of -7.7 to -8.1 kcal may be considered as promising results (Wang et al. 2017; Rehman et al. 2019).
Table 1. Physio-chemical Properties of ligands with Lipinski rule of 5* (Molecular mass less than 500 Dalton, High lipophilicity (expressed as LogP less than 5), Less than 5 hydrogen bond donors, less than 10 hydrogen bond acceptors, Molar refractivity should be between 40-130).
| Ligands
Properties |
Prangenidin | Columbin |
| Chemical Formula | C16H14O4 | C20H22O6 |
| Source | Angelica dahurica | Tinospora cordifolila |
| pH | 3.1 | 1.3 |
| Molecular Weight | 270.28 g/mol
|
358.4 g/mol |
| Hydrogen Bond Donor | 1 | 1 |
| HydrogenBond Acceptors | 4 | 6 |
| LogP | 3.7 | 2.2 |
Table 2. The table shows interaction between the active poles of the
ligand and the amino acid of the receptor (α-glucosidase).
| Receptor | Ligand | Amino acid interacted | Distance between the amino acid and the ligand pole (Å) |
| α-glucosidase | Prangenidin | ASP-202 | 2.8 |
| α-glucosidase | Prangenidin | GLU-271 | 2.3 |
Figure 1: (a) The docked pose of ligand (prangenidin) in the binding pocket of the enzyme which is used as a receptor; α-glucosidase. The ligand is shown in the stick structure and the enzyme is shown in cartoon model structure. (b) This figure depicts the H-bond interaction between the ligand, prangenidin with the residues of α-glucosidase which are GLU-271 and ASN-202. The blue dots show the H-bonding between the ligand and the residue. The numbers with the H-bonds represents the distance between the residue and the ligand interacting pole (Source: PDB,
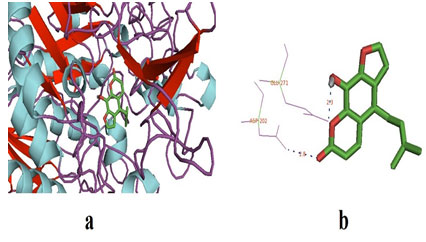
Table 3. This table shows the receptor and the ligand docked, force field with their
binding affinities at different position and the best energy minimization being -7.7.
| Ligand | Binding Affinity | rmsd/ub | rmsd/lb |
| a-glucosidase_69502 | -7.7 | 0 | 0 |
| a-glucosidase_69502 | -7.2 | 10.031 | 6.86 |
| a-glucosidase_69502 | -7.2 | 11.653 | 9.153 |
| a-glucosidase_69502 | -7 | 11.202 | 9.483 |
| a-glucosidase_69502 | -7 | 16.273 | 14.211 |
| a-glucosidase_69502 | -6.7 | 12.724 | 10.256 |
| a-glucosidase_69502 | -6.7 | 16.999 | 13.74 |
| a-glucosidase_69502 | -6.7 | 12.748 | 9.634 |
| a-glucosidase_69502 | -6.6 | 12.966 | 9.359 |
PyRx & BIOVIA).
Table 4. This table shows the receptor and the ligand docked, force field with their
binding affinities at different position and the best energy minimization being -8.1.
| Ligand | Binding Affinity | rmsd/ub | rmsd/lb |
| a-glucosidase_442015 | -8.1 | 0 | 0 |
| a-glucosidase_442015 | -7.3 | 3.166 | 2.054 |
| a-glucosidase_442015 | -7.3 | 6.81 | 2.048 |
| a-glucosidase_442015 | -7.3 | 34.481 | 31.079 |
| a-glucosidase_442015 | -7.3 | 34.359 | 32.715 |
| a-glucosidase_442015 | -6.8 | 21.467 | 19.651 |
| a-glucosidase_442015 | -6.8 | 6.676 | 3.846 |
| a-glucosidase_442015 | -6.6 | 34.255 | 31.272 |
| a-glucosidase_442015 | -6.4 | 33.31 | 31.553 |
Figure 2: (a) The docked pose of ligand (Columbin) in the binding pocket of the enzyme which is used as the receptor; α-glucosidase. The ligand is shown in the stick structure and the enzyme is shown in cartoon model structure. (b). This figure depicts the H-bond interaction between the ligand, columbin with the residues of α-glucosidase which is ASN-301. The blue dots show the H-bonding between the ligand and the residue. The number with the H-bonds represents the distance between the residue and the ligand interacting pole (Source: PDB, PyRx & BIOVIA).
In discussion, there have been a variety of pharmacologically active metabolites reported from plant sources. Evidences have suggested that these bioactive metabolites are majorly a part of our diet and demonstrated their role in the treatment of several dreadful diseases including cardiovascular, cancer and neruro-degeneration (Kumar et al. 2015; Kashyap et al. 2016; Kashyap et al. 2018a; Kashyap et al. 2018b; Aggarwal et al. 2019; Kashyap et al. 2019; Tuli et al. 2019; Sharma et al. 2019).
Therefore, diet modification may contribute to preventing a significant number of such dreadful diseases. Furthermore, existing conventional approaches of drug discovery are very much time consuming and labor-oriented. Moreover, diagnosis and progression rate of such diseases are very fast. Therefore, in such a drastic scenario computational biology may help the pharmaceutical industry to fast the drug discovery processes (Wooller et al. 2017).
Computational tools of docking can not only tell the interactions of the ligand with receptor but also define the possibility of drug synthesis from a large number of chemical database (Hasselgren and Myatt 2018; Thomford et al. 2018; Mochizuki et al. 2019; Patel et al. 2020; Gupta et al. 2021). Therefore in order to understand the interactions and binding affinities of ligands with glucosidase receptor, we performed molecular docking studies by using PyRx software. Out of tested ligand columbin was found to interact more firmly with glucosidase receptor with more docking score as well as H-bond formation capability.
Furthermore results of present study explore the binding affinity of both the ligands for glucosidase receptor with active interacting amino acid residues GLU 271, ASP 202, and ASN 301. Previously also in silico studies of phytochemicals or their synthetic derivatives such as oriciacridone F and O-methylmahanineand 2-(benzo[d][1,3]dioxol-5-yl)-4H-chromen-4-one, respectively have been carried to find α-Glucosidase Inhibitors (Zafar et al. 2016; Meena et al. 2019).
More recently, three compounds named as 50-hydroxymethyl-10-(1, 2, 3, 9-tetrahydro- pyrrolo(2,1-b)quinazolin-1-yl)-heptan-10-one(1),-terpinyl-glucoside (2), and machaeridiol-A were extracted from Psychotria malayana and docked with glucosidase (Nipun et al. 2020). Results revealed that four hydrogen bonds formed at ASP352, ARG213, ARG442, GLU277, GLN279, HIE280, and GLU411 and energy minimizarion were in the range of -7.6, and -10.0 kcal/mole. Therefore results of present study are in good agreement with previously published work so as to find out active interacting residue as well binding affinities.
CONCLUSION
The findings of the present study revealed molecular docking between ligands (Prangenidin and Columbin) and α-glucosidase. We found that the binding energies of these two ligands ranging from -6.4 to -8.1 and also the hydrogen bond interaction are quite strong. Diabetes mellitus is one of the biggest threats to human health which is increasing at an alarming rate. This work is an effort to put forward these two ligands as future anti-hyperglycemic agents.
Conflict of interest: Authors declare no conflicts of interests to disclose.
REFERENCES
Abdelwahab SI, Koko WS, Taha MME et al. (2012). In vitro and in vivo anti-inflammatory activities of columbin through the inhibition of cycloxygenase-2 and nitric oxide but not the suppression of NF-κB translocation. Eur J Pharmacol 678:61-70. doi: 10.1016/j.ejphar.2011.12.024.PMID: 22227329.
Aggarwal V, Kashyap D, Sak K et al. (2019). Molecular Mechanisms of Action of Tocotrienols in Cancer: Recent Trends and Advancements. Int J Mol Sci 20:656. doi: 10.3390/ijms20030656.PMID: 30717416.
Campbell LK, Baker DE, and Campbell RK (2000). Miglitol: Assessment of its role in the treatment of patients with Diabetes mellitus. Ann Pharmacother 34:1291–1301.doi: 10.1345/aph.19269.PMID: 11098345.
Chang J, Block TM and Guo JT (2013). Antiviral therapies targeting host ER alpha-glucosidases: current status and future directions. Antiviral Res 99:251-60. doi: 10.1016/j.antiviral.2013.06.011.PMID: 23816430a
Chen Q, Li P, He J et al. (2008). Supercritical fluid extraction for identification and determination of volatile metabolites from Angelica dahurica by GC-MS. J Sep Sci 18:3218-24.doi: 10.1002/jssc.200800325.PMID: 18705001.
Chen TH, Fu YS, Chen SP et al. (2021). Garcinia linii extracts exert the mediation of anti-diabetic molecular targets on anti-hyperglycemia. Biomed Pharmacothe 134: 111151. doi.org/10.1016/j.biopha.2020.111151
Chung S, Kim S, Son M, et al. (2019). Empagliflozin contributes to polyuria via regulation of sodium transporters and water channels in diabetic rat kidneys. Front Physiol 10: 271. doi: 10.3389/fphys.2019.00271.
Gamblin DP, Scanlan EM and Davis BG (2009). Glycoprotein synthesis: An update. Chem Rev 109 : 131-163. https://doi.org/10.1021/cr078291i
Gupta R, Srivastava D, Sahu M et al. (2021). Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol Divers 1-46. doi: 10.1007/s11030-021-10217-3
Hasselgren C, and Myatt GJ (2018). Computational Toxicology and Drug Discovery. Methods Mol Biol 1800:233-244. doi: 10.1007/978-1-4939-7899-1_11.PMID: 29934896.
Hsiao SH, Liao LH, Cheng PN et al. (2006). Hepatotoxicity associated with acarbose therapy. Ann. Pharmacother 40: 151–154.doi: 10.1345/aph.1G336.PMID: 16317107.
Humphries MJ, Matsumoto K and White SL (1986). Inhibition of experimental metastasis by Castanospermine in mice: blockage of two distinct stages of tumor colonization by oligosaccharide processing inhibitors. Cancer Res 46: 5215-5222. PMID: 3093061.
Hwangseo P, Kyo YH, Kyung H et al. (2008). Discovery of novel α-glucosidase inhibitors based on the virtual screening with the homology-modelled protein structure. Bioorg Med Chem 16: 284-292.doi: 10.1016/j.bmc.2007.09.036.PMID: 17920282.
Kashyap D, Sharma A, Tuli HS et al. (2018a) Molecular Targets of Celastrol in Cancer: Recent Trends and Advancements. Crit Rev Oncol Hemato 128:70-81. doi: 10.1016/j.critrevonc.2018.05.019.PMID: 29958633.
Kashyap D, Mondal R, Tuli HS et al. (2016). Molecular Targets of Gambogic Acid in Cancer: Recent Trends and Advancements. Tumour Biol 3710:12915-12925. doi: 10.1007/s13277-016-5194-8.PMID: 27448303.
Kashyap D, Sharma A, Sak K et al. (2018b). Fisetin: A Bioactive Phytochemical With Potential for Cancer Prevention and Pharmacotherapy. Life Sci 1:75-87.doi: 10.1016/j.lfs.2017.12.005. PMID: 29225112
Kashyap D, Tuli HS, Yerer MB et al. (2019). Natural Product-Based Nanoformulations for Cancer Therapy: Opportunities and Challenges. Seminar Cancer Biol 14: S1044-579X(19)30103-8. doi: 10.1016/j.semcancer.2019.08.014.PMID: 31421264.
Kato-Schwartz CG, Corrêa RCG, Souza LDD et al. (2020). Potential anti-diabetic properties of Merlot grape pomace extract: An in vitro, in silico and in vivo study of α-amylase and α-glucosidase inhibition. Food Res Int 137: 109462. doi: 10.1016/j.foodres.2020.109462
Krentz AJ and Bailey CJ (2005). Oral antidiabetic agents: current role in type 2 Diabetes mellitus. Drugs 65:385–411. doi: 10.2165/00003495-200565030-00005.PMID: 15669880.
Kumar G, Tuli HS, Mittal S et al. (2015). Isothiocyanates: A Class of Bioactive Metabolites With Chemopreventive Potential. Tumour Biol 36:4005-16. doi: 10.1007/s13277-015-3391-5.PMID: 25835976.
Lefebvre PJ and Scheen AJ (1994). The use of acarbose in the prevention and treatment of hypoglycaemia. Eur J Clin Invest 24:40–44. doi:10.1111/j.1365-2362.1994.tb02255.x.PMID: 8001627.
Li H, Chao X, He CL et al. (2016). Alloimperatorin and Its Epoxide Derivative Exhibit In Vitro Antitumor Activity in HL-60 Acute Myeloid Leukemia Cancer Cells via Inducing Apoptosis, Cell Cycle Disruption and Inhibition of Cell Migration. Bangladesh J Pharmacol 11: 194-99.doi: 10.3329/bjp.v11i1.24575.
Mata R, Cristians S and Escandon R (2013). Mexican anti-diabetic herbs: valuable source of inhibitor of alpha glucosidases. J Nat Prod 76: 468-483.doi: 10.1021/np300869g.PMID: 23398496.
Meena SN, Kumar U, Naik MM et al. (2019). α-Glucosidase Inhibition Activity and in Silico Study of 2-(benzo[d][1,3]dioxol-5-yl)-4H-chromen-4-one, a Synthetic Derivative of Flavone. Bioorg Med Chem 27: 2340-2344. doi: 10.1016/j.bmc.2018.12.021.PMID: 30594450
Mochizuki M, Suzuki SD, Yanagisawa K et al. (2019). QEX: target-specific druglikeness filter enhances ligand-based virtual screening. Mol divers 23(1), 11-18. doi: 10.1007/s11030-018-9842-3.
Nathan, DM, Buse JB, Davidson MB et al. (2006). Management of hyperglycaemia in type 2 diabetes: A consensus algorithm for the initiation and adjustment of therapy. A consensus statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetologia 49: 1711–1721.doi: 10.2337/dc06-9912.PMID: 16873813.
Nipun TS, Khatib A, Ibrahim Z et al. (2020). Characterization of α-Glucosidase Inhibitors from Psychotria malayana Jack Leaves Extract Using LC-MS-Based Multivariate Data Analysis and In-Silico Molecular Docking. Molecules 25: 5885. doi: 10.3390/molecules25245885
O’Keefe S, Roebuck QP, Nakagome I et al. (2019). Characterizing the selectivity of ER α-glucosidase inhibitors. Glycobiology 29: 530-542. doi: 10.1093/glycob/cwz029.
Park H, Hwang KY, Kim YH et al. (2008). Discovery and biological evaluation of novel α-glucosidase inhibitors with in vivo antidiabetic effect. Bioorg Med Chem Lett 18: 3711-3715.doi: 10.1016/j.bmcl.2008.05.056 PMID: 18524587.
Patel L, Shukla T, Huang X et al. (2020). Machine learning methods in drug discovery. Molecules 25: 5277. doi: 10.3390/molecules25225277.
Playford RJ, Ither C and Gao R (2013). Use of alpha-glucosidase inhibitor acarbose in patient with ‘Middleton syndrome’, normal gastric anatomy but with accelerated gastric emptying causing postprandial reactive hypoglycemia and diarrhea. Can J Gastroentrol 27: 403-404.doi: 10.1155/2013/791803.PMID: 23862171.
Raghu C, Arjun HA and Anantharaman P (2019). In vitro and in silico inhibition properties of fucoidan against α-amylase and α-D-glucosidase with relevance to type 2 diabetes mellitus. Carbohydr Polym 209, 350-355. doi: 10.1016/j.carbpol.2019.01.039
Rawling AJ, Lomas H, Adam WP et al. (2009). Synthesis and biological characterization of novel N-alkyl deoxynojirimycin α-glucosidase inhibitors. Chem Bio Chem 10: 1101-1105.doi: 10.1002/cbic.200900025.PMID: 19294724.
Rudd PM, Storr SJ, Royle L et al. (2018). The O-linked glycosylation of secretory/shed MUC1 from an advanced breast cancer patient’s serum. Glycobiol 18:456-462.doi: 10.1093/glycob/cwn022.PMID: 18332077.
Scott LJ and Spencer CM (2000). Miglitol: A review of its therapeutic potential in type 2 Diabetes mellitus. Drugs 59: 521–549. doi: 10.2165/00003495-200059030-00012.PMID: 10776834.
Sharma A, Ghani A, Sak K et al. (2019). Probing Into Therapeutic Anti-cancer Potential of Apigenin: Recent Trends and Future Directions. Recent Pat Inflamm Allergy Drug Discov 13:124-133.doi: 10.2174/1872213X13666190816160240.PMID: 31418666
Thomford NE, Senthebane DA, Rowe A et al. (2018). Natural products for drug discovery in the 21st century: innovations for novel drug discovery. Int J Mol Sci 19:1578. doi: 10.3390/ijms19061578
Tuli HS, Sood S, Bhatia GK et al. (2021). In silico analysis and molecular docking studies of plumbagin and piperine ligands as potential inhibitors of alphaglucosidase receptor. Biointerface Res Appl Chem 11:9629-9637. doi.org/10.33263/BRIAC0112.85878598
Tuli HS, Tuorkey MJ, Thakral F et al. (2019). Molecular Mechanisms of Action of Genistein in Cancer: Recent Advances. Front Pharmacol 10:1336. doi: 10.3389/fphar.2019.01336. PMID: 31866857.
Rehman NU, Rafiq K, Khan A et al. (2019). α-Glucosidase inhibition and molecular docking studies of natural brominated metabolites from marine macro brown alga Dictyopteris hoytii. Mar drugs 17:666. doi: 10.3390/md17120666.
Venable JS and Aschenbrenner SD (2002). Drug Therapy In Nursing. Hagerstown, MD: Lippincott Williams & Wilkins. Wolters Kluwer Health Adis (ESP). 1-1346.
Wang GC, Peng YP, Xie ZZ et al. (2017). Synthesis, α-glucosidase inhibition and molecular docking studies of novel thiazolidine-2, 4-dione or rhodanine derivatives. Med Chem Comm 8:1477-84. doi: 10.1039/c7md00173h.
Wooller SK, Hume GM, Chen X et al. (2017). Bioinformatics in Translational Drug Discovery. Biosci Rep 37:BSR20160180. doi: 10.1042/BSR20160180.PMID: 28487472.
Yin Z, Zhang W, Feng F et al. (2014). α-Glucosidase inhibitors isolated from medicinal plants. Food Sci Human Wellness 3:136–174. doi.org/10.1016/j.fshw.2014.11.003.
Yousuf H, Shamim S, Khan KM et al. (2020). Dihydropyridines as potential α-amylase and α-glucosidase inhibitors: synthesis, in vitro and in silico studies. Bioorg chem 96, 103581. doi: 10.1016/j.bioorg.2020.103581
Zafar M, Khan H, Rauf A et al. (2016). In Silico Study of Alkaloids as α-Glucosidase Inhibitors: Hope for the Discovery of Effective Lead Compounds. Front Endocrinol 7:153. doi: 10.3389/fendo.2016.00153.PMID: 28066324.
Zhang L, Song P, Zhang X et al. (2020). alpha-Glucosidase inhibitors alter gut microbiota and ameliorate collagen-induced arthritis. Front Pharmacol 10: 1684. doi: 10.3389/fphar.2019.01684.


