Laboratory of Food Chemistry and Microbiology,Food & Nutrition Division
Department of Home Science. University of Calcutta. Kolkata-700027.
Corresponding author email: soumyanandi1992@gmail.com
Article Publishing History
Received: 24/03/2024
Accepted After Revision: 28/05/2024
Introduction: Looking for novel amylases needed for industrial processes may benefit from the screening of microorganisms with high amylase activity. Microbial enzymes are widely used in industrial processes due to affordable cost, significant production, chemical stability, environmental protection, plasticity, and widespread availability. Since Purba Bardhaman contains numerous rice processing plants and agro-industrial waste disposal locations, water samples were gathered for the study. Material & Methods: Amylase-producing bacteria were isolated, identified and revealed from this work to add new knowledge into the field of science.
Results: Seven bacterial isolates were isolated, and based on appearance of zone of hydrolysis in starch agar plates was selected for further study. The isolate was Gram-positive, spore-forming rods. Depending on 16s rRNA gene sequences with 99.79% similarity the isolate was identified as Brevibacillus parabrevis, efficient amylase producers with an excellent yield throughout the solid-state fermentation technique. The isolate demonstrated maximum enzyme activity of 350 U/mg at 5% substrate concentrations. This piece of work focuses on finding out whether agro-industrial waste water could be a better source for amylolytic bacteria.
Agro-Industrial Waste Water, Amylolytic Bacteria, Biochemical Characterization, Screening.
Nandi S, Chatterjee A. Identification and Characterization of Amylolytic Bacteria from Agro-industrial Waste Water. Biosc.Biotech.Res.Comm. 2024;17(2).
Nandi S, Chatterjee A. Identification and Characterization of Amylolytic Bacteria from Agro-industrial Waste Water. Biosc.Biotech.Res.Comm. 2024;17(2).Available from: <a href=”https://shorturl.at/b0u8y“>https://shorturl.at/b0u8y</a>
INTRODUCTION
Microbial population can be manipulated to yield enzymes which are commercially important in organic compound synthesis, clinical analysis, pharmaceuticals, detergents, food production and fermentation. Microorganisms can easily be targeted as economical source for industrial enzyme production. Use of cheap, easily accessible wastes, such as agro-industrial waste, as a novel substrate for production and synthesis of amylase for industrial use is an ongoing effort that helps to address pollution issues. Amylase, an enzyme, is needed for the catalytic degradation of starch into its monomeric elements, of which glucose is the smallest, (Logeswaran et al.2014, Saha et al.2019).
Looking for novel amylases needed for industrial processes may benefit from the screening of other microorganisms with high amylase activity. Microbial production of amylase is more fruitful than other sources like plants or animals, because of short growth period, biochemical diversity and simplicity with environmental and genetic manipulation could improve the capacity for enzyme synthesis, Mishra and Behera (2008), Saha et al (2019). In order to substitute enzymes, which are typically extracted from complex eukaryotes because to their biochemical diversity and the ease with which enzyme concentrations may be increased by environmental and genetic manipulation, attempts are now being explored, (Bole et al.2013). Microbial enzymes are widely used in industrial processes due to affordable cost, significant production, chemical stability, environmental protection, plasticity, and widespread availability, (Mishra and Behera 2008, Deb et al.2013, Burhan et al.2003).
Approximately 25% of the enzyme market is comprised of amylase enzyme. Fungi and bacteria are the best choices of the source because they are very economical with high production rate and can be genetically engineered for the desired quality and quantity of amylase production, (Islam et al 2017). In biotechnological applications ranging from food fermentation, detergent, pharmaceutical, brewing, and textile to paper industries, amylases perform a significant role, (Kathiresan and Manivannan 2006). Low-cost amylase production is necessary to meet the greater demands of these sectors, (Saxena and Singh 2011). Earlier, amylase production has been studied using submerged (SmF) and solid-state fermentation (SSF), Perez-Guarre et al. (2003), Saxena and Singh (2011).
However, as the elements of a synthetic medium are very costly and uneconomical, they must be substituted with agricultural and industrial waste products, which are thought to be a viable source for microbial populations that produce enzymes and are more readily available. In this present experimental work, isolation and screening of amylase producing microbes has been attempted with samples collected from rice processing units in Bardhhaman, West Bengal. Since these areas are rich in rice processing units and agro-industrial waste, therefore, data of this work might contribute new insights into finding cheaper sources for amylase producing microorganisms. This process was achieved by stepwise activities including isolation of α-amylase producing microorganisms from the waste water sources; selection and identification of the most potent isolate for in-vitro enzyme production while utilizing the solid media; and optimization of culture conditions affecting α-amylase production by the selected isolates.
MATERIAL AND METHODS
Isolation of microorganism: Waste water sample was collected from agro-industrial dump of rice processing unit of Purba Bardhaman, West Bengal India. The study region is situated at 23°15’15.3″N, 88°01’50.9″E latitude and longitude. Water sample was collected by using sterile containers and was stored in 4ºC for subsequent analysis. As the water sample was collected from the dump area of rice processing unit, there might be presence of amylolytic microorganisms so selective media along with basic media was used for isolation of microbes. 0.1 mL of every specimen has been put into nutrient agar plates from the container. (Beef extract 10g/L; Peptone 10 g/L; Sodium Chloride 5 g/L; Agar 15 g/L; pH 7.2±0.1) as well as starch agar plates (Beef extract 3 g/L; Peptone 5 g/L; Soluble starch 10 g/L; Agar 15 g/L; pH 7.2 ± 0.1) in triplicate. After that, the microbial culture was spread using spreader and maintained for 24 hours at 37°C. Colonies were cultured on the proper medium after the incubation time to generate pure isolates, and were then kept at 4°C for more study.
Screening of microorganism: The pure microbial culture obtained was cultured on starch agar plates to authenticate whether the microbial culture obtained was amylolytic bacterial species. After 24 hours of incubation, the plates were immersed with iodine solution for 30 seconds. Clear zones encircling the growth of microbes were considered to be amylase producers.
Characterization and identification of amylase producing isolates: The isolates were assessed by the gram reaction and colony morphology, respectively using the methods of Collins and Lyne. Collins et al.(2004), Sinha.(2010). Additionally, other biochemical assays such as IMViC Test, Urease test, and Starch hydrolysis tests, sugar fermentation, nitrate reduction test were from carried out to characterize isolates in terms of their biochemistry. As the sample was collected from waste water so coliform test was also carried out. 16s rRNA gene sequences was also carried out for authentication of the species and genera of the microbes. Saitou and Nei, (1987).
Standardization of substrate concentration and incubation period for enzyme production: Substrate concentration for enzyme production was optimized using different concentrations of starch (1.0, 2.5, 5.0, 7.5 and 10.0%) with different incubation period of time (24, 48, 72, 96 and 120 h) in the production medium.
Amylase production by using Solid State Fermentation (SSF): After standardization of the substrate concentration and incubation period, the SSF process was carried out using rice husk. Substrate bed (Rice Husk: 10 g, Starch: 0.5 g, KH2PO4: 0.2 g, NaCl: 0.25 g, MgSO4, 7H2O: 0.02 g, CaCl2:0.1 g, (NH4)2SO4: 0.1 g) was prepared and transferred into 250 mL capacity Erlenmeyer flasks. Moisture content was adjusted with 20 mL de-ionized water and autoclaved and allowed to cool to room temperature. 10 mL of 24 h old microbial cultures was added into the substrate medium and incubated for 96 h, (Tsegaye et. al.2014).
Extraction of crude enzyme: The extracellular enzymes from the fermented substrate were extracted using phosphate buffered saline (50 mL) after proper agitation on a rotary shaker at 120 rpm for 45 minutes. The content was filtered and squeezed through a cotton cloth. The filtrate was used as the crude enzyme, Tsegaye et. al. (2014).Determination of enzyme activity: Activity of α-amylase was determined by 3,5 Di-nitro salicylic acid (DNSA) method, Miller (1959) using Potato Starch as substrate and Sodium Phosphate buffer (pH-6.9) as the incubation medium at 37°C of incubation temperature. Total protein content was determined, Lowry et al (1951).
RESULTS AND DISCUSSION
Isolation & Screening of Microorganism: As a result of the preliminary screening, many isolates having the capacity to synthesize amylase at different levels were identified. Seven potential microbial isolates were obtained from the waste water samples were marked as Sp1-Sp7 depending upon their growth on nutrient agar as well as clear zone formation in starch agar media after reacting with gram’s iodine solution (Table-01) and this outcome was consistent with the findings of Hmidet et. al. (2009) from the starch hydrolysis test, Sp1 was selected for further investigation.
Table 1. Bacterial Isolates with their clear zone on Starch agar
| Bacterial Isolates | Clear Zone(mm) |
| Sp1 | 25.0 ± 0.5 |
| Sp2 | 10.0 ± 0.1 |
| Sp3 | 17.0 ± 0.2 |
| Sp4 | 15.0 ± 0.6 |
| Sp5 | 8.0 ± 0.1 |
| Sp6 | 4.0 ± 0.9 |
| Sp7 | 1.5 ± 0.3 |
Characterization of amylase producing isolates:
Colony Morphology: The most potent isolate that showed highest starch hydrolyzing ability was selected for further characterization. The isolate was characterized based on colony growth feature and microscopic observation (magnification of 100X and 400X) to distinguish their respective genera. Sp1 demonstrated a regular form, with color and rod shape of colony morphology (Table-02).
Table 2. Colony morphology of the Isolates
| Characteristics | Bacterial Species (Sp1) |
| Configuration | Round |
| Margin | Entire |
| Elevation | Raised |
| Surface | Smooth |
| Density | Opaque |
| Pigmentation | White |
| Gram reaction | Positive (+ Ve) |
| Cell morphology and spore | Rod shaped and Oval |
Effect of different physico-chemical factors: Effect of different physico-chemical factors like temperature, pH and salt concentration (NaCl) was also observed. As per observation the microbial culture is adaptable to a vast range of temperature from 20ºC-45ºC with optimum growth at 37ºC. The isolated microbes are capable to grow even in lower salt concentration (2%) even at lower water activity i.e. xerophile in nature. Biochemical Characterization: Based on the biochemical characterization (Table-03) isolate may be Brevibacillus sp. Depending on the isolate’s initial screening, these genera might be suitable for commercial applications, which the previous report corroborated by Ashwini et al. (2011).
Table 3. Biochemical Characteristics of Bacterial Isolate
| Biochemical tests | Bacterial Isolate (Sp1) | |
| Growth on Mac Conkey agar medium | – | |
| Indole test | + | |
| Methyl red test | – | |
| Voges Proskauer test | – | |
| Citrate hydrolysis | – | |
| Casein hydrolysis | – | |
| Starch hydrolysis | + | |
| Gelatin hydrolysis | – | |
| Nitrate reduction | – | |
| Catalase | – | |
| Esculine hydrolysis | + | |
| H2S gas production | – | |
|
Acid production from carbohydrates |
Dextrose | – |
| Fructose | – | |
| Sucrose | – | |
| Lactose | – | |
+ Positive: – Negative
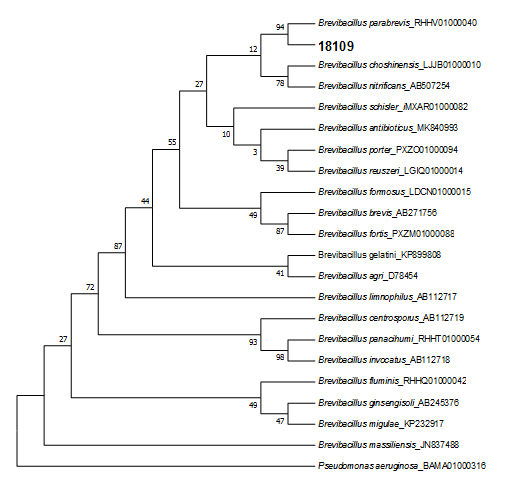
16s rRNA gene Sequencing: The best amylase producing bacteria was isolated and identified by amplification and sequencing of its 16s rRNA full length coding gene, following the comparison of the obtained sequence with the NCBI database using the BLAST tool. The result showed that the selected stain was closely to Brevibacillus gene and in particular to the species parabrevis with 99.79% similarity. Therefore, the newly isolated strain was named as Brevibacillus parabrevis 18109.
Figure 1: Phylogenetic tree of strain 18109 based on 16S rRNA gene sequences.
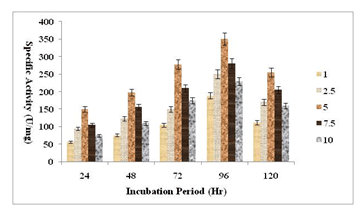
Standardization of substrate concentration & Incubation period for enzyme production: Fermentation conditions need to be optimized, especially with regard to physical and chemical characteristics, Wenster-Botz (2000). Amylase activity generally increased while starch concentration increased from 1.0% to 10.0%. In this investigation, 5% of starch content provided the maximum activity (Figure-02) at different incubation time. The present finding is also endorsed with previous investigations on amylase activity, as reported by Oyeleke and Oudwole (2009). At 96 hours into the fermentation procedure, the isolate’s amylase activity reached its peak, after which it began to drop (Figure-02). Bacillus subtilis and Bacillus sp. DLB9 has also exhibited comparable outcomes, Shyam et al.(2013).
Figure 2: Standardization of Substrate Concentration & incubation Period
Determination of potential enzyme activity: The crude enzyme isolated from Sp1 showed the maximum specific activity of 350 U/mg, the current results are consistent with past reports obtained from Bacillus species, Bukhari and Rehman (2015).
The experimental piece of work was focused on obtaining microbial isolates from agro-industrial waste water having the ability to produce amylase. Many microbial species have already been identified as good amylase producers. Studies of amylases from bacteria and fungi are well available but using a cheaper resource for microbial isolation has not been documented. In our study, seven microbial isolates were identified from the five different waste water samples collected from the rice processing units. Though most of the isolated microorganisms are potent producer amylase but depending upon the starch breaking capacity of amylase produced by the microorganisms on starch agar media, Sp1 was selected for further investigation. Being neutrophilc in nature the microbe is adaptable to live in an environment where the hydrogen ion concentration is at equilibrium i.e. thrives in a relatively neutral pH, in the range of pH 5-9.
Even in lower salt concentration, the isolated microbe is capable to grow, which indicates its xerophilic nature. With 99.79% similarity, the isolated microbial stain was closely related to the Brevibacillus genus and in particular to the species parabrevis, potential candidates with several industrial applications for amylase production. Generally, the amount of amylase activity increased as the starch concentration rose from 1.0% to 10.0% with maximum activity at 5% concentration, but when excludes 5% concentration, amylase activity decreased. This might be because the isolates have the ability to metabolize starch within a short amount of time after the concentration was raised. The isolate showed maximum amylase activity at 96 h. The suppression and presence of other byproducts in the fermentation medium as well as a reduction in nutrients may be the causes of the amylase activity decline after 96 hours, Haq et al (2010), Gebreyohannes (2015).
CONCLUSION
The present study attempts to explore the potential of indigenously microbial isolates from easily available cheap source that are capable to produce amylases. The current findings bring about a decision that the agro-industrial waste water possesses the potential to be a source of amylase-producing microbes that might be used to create highly effective industrial amylases. The present study is the first-time report on the capability of alpha amylase producing activity by Brevibacillus parabrevis. The isolated bacterial species that showed higher amylase activity can be characterized and exploited further for various useful industrial applications.
REFERENCES
Ashwini K., Gaurav K., Karthik L., Bhaskara Rao K.V. (2011). Optimization, production and partial purification of extracellular α-amylase from Bacillus sp. marini. Arch. Appl. Sci. Res. Vol 3 No 1 Pages 33-42.
Bole S., Maji A., Dey A., Acharya A., Dubey S., Lal R. (2013). Isolation, Purification and Characterization of amylase from airborne-bacteria. World journal of pharmacy and pharmaceutical sciences. Vol 2 No 6.
Bukhari D.A., Rehman A. (2015). Purification and Characterization of α-Amylase from Bacillus subtilis Isolated from Local Environment. Pakistan J. Zool. Vol 47 No 4 Pages 905-911.
Burhan A., Nisa U., Gokhan C., Omer C., Ashabil A., Osman G. (2003). Enzymatic properties of a novel thermophilic, alkaline and chelator resistant amylase from an alkalophilic Bacillus sp. Isolate ANT-6. Process Biochem. Vol 38 Pages 1397–1403
Collins C.H., Lyne P.M., Grange J.M., and Falkinham J.O. (2004). Collins and Lyne ‘s Microbiological Methods: Arnold, a member of the Hodder Headline Group.
Deb P., Talukdar S.A., Mohsina K., Sarker P.K. and Abu S.M. (2013). Production and partial characterization of extracellular amylase enzyme from Bacillus amyloliquefaciens P-001. Springer Plus. Vol 2 No 154
Gebreyohannes G. (2015). Isolation and optimization of amylase producing bacteria and actinomycetes from soil samples of Maraki and Tewedros campus, University of Gondar, North West Ethiopia. African Journal of Microbiology Research. Vol 9 No 31 Pages 1877-1882.
Haq H., Ashaf M.A., Qadeer J. (2010). Pearl millet, a source of α-amylase production by Bacillus licheniformis. Bioresour. Technol. Vol 96 Pages 1201-1204.
Hmidet N., Hadj Ali N.,Haddar A., Kanoun S, Alya S, Nasri M. (2009). Alkaline proteases and thermostable alpha-amylase co-produced by Bacillus licheniformis NH1: Characterization and potential application as detergent additive. Biochem. Eng. J.Vol 47 Pages 71-79.
Islam T., Choudury N., Mahboob Hossain M., Khan T.T. (2017). Isolation of Amylase Producing Bacteria from Soil, Identification by 16S rRNA Gene Sequencing and Characterization of Amylase. Bangladesh J Microbiol. Vol 34 No 1 Pages 01-06
Kathiresan K., Manivannan S. (2006). α Amylase production by Penicillium fellutanum isolated from mangrove rhizosphere soil. African J. Biotech. Vol 5 No 10 Pages 829-832.
Logeswaran R., Prabagaran S.R.P. Ramesh D. (2014). Bacterial diversity towards industrially important enzyme producers from Velliangiri Hills, Western Ghats. J. Env. Sci. Toxicol. Food Tech. Vol 8 No 5 Pages 45-63.
Lowry O.H., Rosenbrough N.J., FarrA.L. and Randall R.J. (1951). Protein Measurement with the Folin Phenol Reagent. J. Bio Chem. Vol 193 Pages 265-275
Miller, G. (1959). Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Analytical Chemistry. Vol 31 Pages 426-428
Mishra S and Behera N. (2008). Amylase activity of a starch degrading bacteria isolated from soil receiving kitchen wastes. Afr. J. Biotechnol. Vol 7 No 18 Pages 3326-3331.
Oyeleke S.B., Oduwole A.A. (2009). Production of amylase by bacteria isolated from a cassava waste dump site in Minna, Niger State, Nigeria. Afr. J. Microbiol. Res. Vol 3 No 4 Pages 143-146.
Perez-Guarre N., Torrado-Agrasar A., Lopez-Macias C., Pastrana L. (2003). Main characteristics and application of solid substrate fermentation, Electron. J. Environ. Agric. Food Chem. 2:243–350.
Saha M.L., Nowshin K.I, AkterT.,Rahman I.A., IslamT.andKhan T. (2019). Isolationand identification of amylolytic bacteria from garbage and garden soil. Bangladesh J. Bot. Vol 48 No 3 Pages 537-545,
Saitou N. and Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. Vol 4 Pages 406-425.
Saxena R., Singh R. (2011). Amylase Production by Solid-state Fermentation of Agro-Industrial Wastes using Bacillus sp. Brazilian Journal of Microbiology. Vol 42 Pages 1334-1342
ShyamS.A., Sonia S.S., Lal G. (2013). Amylase activity of a starch degrading bacteria isolated from soil. Arch. App. Sci. Res. Vol 5 No 1 Pages 15- 24.
Sinha, C. (2010). Isolation, Characterization and Optimization of amylase producing bacteria from Municipal waste. Ph.D thesis, Jadavpur University, India
Tsegaye K. N. and Gessesse A. (2014). Amylase Production under solid state fermentation by a bacterial isolate W74. African Journal of Biotechnology. Vol 13 No 21 Pages 2145-2153
Wenster-Botz D. (2000). Experimental design for fermentation media development: Statistical design or Global random search? J. Biosci. Bioeng. Vol 90 No 5 Pages 473–483.


