*BS Patel College, Pimpalgaon Kale
**Postgraduate Department of Zoology, Shri Shivaji College of Science Akola
#RLT College of Science Akola MS India
Corresponding author email: poojaguhilot303@gmail.com
Article Publishing History
Received: 07/10/2019
Accepted After Revision: 08/12/2019
DNA from seven isolates of ixodid ticks was collected from the livestock in and around Akola District Maharashtra India, and was analyzed by RAPD using PCR. Selected three casual primers were used for the study of genetic analysis among different isolates of ixodid ticks. A high degree of genetic polymorphism with a unlike pattern of RAPD profiles for each tick isolate was identified with all these random primers. This variability was also established by similarity coefficient values and dendrogram which were perform using mean RAPD profiles for all the primers between different isolates of ticks. The conclusion suggest existence of a complex genotypic diversity of the ixodid tick in Akola district, first time.
Ticks, Random primers, RAPD, Genetic variability of ixodid tick, Akola.
Thakur P. S, Raja I. A, Joshi R. P. Genetic Analysis of Ticks from Livestock of Akola District Maharashtra India. Biosc.Biotech.Res.Comm. 2019;12(4).
Thakur P. S, Raja I. A, Joshi R. P. Genetic Analysis of Ticks from Livestock of Akola District Maharashtra India. Biosc.Biotech.Res.Comm. 2019;12(4). Available from: https://bit.ly/2pEVwpC
INTRODUCTION
India is house for approximately 109 tick species, under 12 genera (Ghosh et al., 2007)… The larva, nymph and adult stages are all parasitic as they suck on blood and lymph of their hosts and thus are the vectors of many kinds of pathogens. A basic awareness on genetic diversity of tick population help in understanding the susceptibility or resistance nature of ticks for effective control as well as production of vaccines (Passos et al., 1999). The molecular techniques or tools that are dramatically advanced the knowledge on the control of ticks and the diagnosis of tick-born diseases (Stiller, 1992). Studies on genomic variations within a species have great impact on understanding the pathogen transmission, the epidemiology of the illness and its control.
Random Amplified Polymorphic DNA (RAPD) fingerprinting allows detecting genetic polymorphism using arbitrary primers (Welsh and McCelland, 1990; Williams et al., 1990). Lan et al., (1996) which offers a rapid and efficient method for generating a new series of DNA markers in animal species, (Mishra et al., 2012, Amany et al., 2019).) The results obtained out of such study shows that the primers are diagnostic and variations in different related species under study (Abdul Anvesh et al., 2018). Morphological misidentification could be reduced significantly after using Multiplex-PCR as the pairs of closely related tick could be appropriately differentiated on gel electrophoresis following PCR (Asadolla et al., 2017).
The present investigation was planned to detect the genetic variability through genetic screening using RAPD marker in the populations of tick species for the first time, from different locations in and around Akola district, of Maharashtra .
MATERIALS and METHODS
Experimental ticks were collected from different livestock’s located in different places of Akola district Maharashtra and were identified using their standard methods as per taxonomic descriptions provided by Walker et al., (2003).
For genetic studies, tissue were prepared and DNA purified from tissue using Promega wizard genomic Kit and Quantified using Nanodrop spectrophotometer (ND 1000, Thermo Corporation, USA) and quality checked on 1% Agarose gel electrophoresis. Gel was visualized using gel documentation system (Bio-Rad Inc., USA). DNA diluted to have 100 ng/µl concentrations and stored at -200C for further use. These DNA samples were processed further for amplification of RAPD. Three RAPD primers, were used which were Pz (CGGCCCGGTA) (Lan et al., 1996) OPJ-01(CCCGGATAA),OPJ-02(CCCGTTGGGA) for the first time in Akola district.RAPD amplification was performed in 25 µl PCR reaction using Kapa biosystems PCR kit. The reaction constitutes 17.8 µl Nuclease free water
2.5 µl 10X PCR buffer,
0.5 µl MgCl2,
2.0 µl 2.5mM DNTPs,
2 µl 10mM primer (PZ and OPJ1) and
0.2 µl taq polymerase (5 units/µl)
and 2 µl of 100ng/ µl of template DNA.
The PCR thermal cycle conditions include an initial denaturation step of 5 min at 95⁰C, 35 cycles of 30 sec at 95⁰C, 40 sec at 36⁰C, and 1 min at 72⁰C, with a final extension at 72⁰C for 10 min. Amplified PCR products were visualized on 2% agarose gel and were photographed under UV illumination with gel documentation system.
RESULTS and DISCUSSION
Three RAPD primers, were used which were Pz (CGGCCCGGTA) (Lan et al., 1996) OPJ-01(CCCGGATAA),OPJ-02(CCCGTTGGGA). The RAPD banding profiles were computed and analyzed based on presence or absence of bands. Molecular size of amplified bands were estimated by comparing with known molecular weight marker (1kb bp Universal DNA ladder, KAPA Biosystems, USA) The RAPD finger printing patterns for adult of Hyalomma a. anatolicum, Hyalomma marginatum issaci, Hyaloma hussaini, R.Boophilus microplus Rh.Annulatus. Rh. Haemaphysaloides, Haemaphysalis bispinosa obtained with Pz primer are shown in figure Plate.1. The pattern of bands obtained for each sample was species specific. All amplifications were repeated twice and only reproducible bands were considered for scoring. No amplifications were found from OPJ2 primer fragments.
According to the Photo plate 1, the gel image under gel-documentation showed the amplification from PZ primers, the same bands were recorded in PT2 (Hylomma. issaci) and PT5(Rh. annulatus) giving the close genetic pattern in both the species. Some similar amplification are specifically observed in PT1 (Rh.Boophilus microplus), PT3 (Hyalomma a. anatolicum), PT4 (Hyalomma hussaini) andPT7 (Hylomma bispinosa). In PT7 Hylomma bispinosa species we found some different amplification from the other ticks species.
The same gel image after amplification with the OPJ1 primer showed that, PT1(Rh. Boophilus microplus), PT2 (Hyalomma marginatum isaaci) and PT5 (Rh.Annulatus) giving maximum similar amplifications proved that these 3 species of ticks are genetically very close to each other for the genetic fragments of OPJ1 primer. Amplification for PT3, PT4, PT6 and PT7 these four species to be observed genetically close banding patterns with OPJ1 primer. In species PT6 and PT7 some more amplification are observed.
Ticks are typically recognized with microscopic and morphometric investigation. But, there are many difficulties linked with accurate tick identification (Poucher et al., 1999), as separate keys must be used for different developmental stages: larvae, nymphet, and female and male adult forms. However, these difficulties could be resolved by using molecular genetic markers-based keys (Poucher et al., 1999). The molecular technique, generate the potential to explore DNA at the individual base pair. This is proved to be a much more straight-way of measuring and determining the genetic variation within and between species (Hoy, 1994).
RAPD methods were utilized to generate genetic linkage map for the I. scapularis ticks (Ullman et al., 2003). Yang et al. (2004) used RAPD methods in exploring genetic distance of 7 species of Ixodidae ticks which all showed their specific DNA band. The study by Mohammed and Enan (2018) too used the molecular method and demonstrated that ticks in the UAE are very similar at the genetic level. Thus, it was concluded that RAPD could identified, species of Ixodidae ticks. Different species may have different annealing sites and thus give different patterns of bands (Williams et al., 1990). Lan et al., (1996) reported that at 12 and 13 bands could obtain for H. asialicum and Boophilus microplus using Pz primer, respectively. In our study, similar results were recorded that from both the primers PZ and OPJ1, annealing at 4000 bp base pair was significantly observed. There were not many reports available with reference to ticks population and genetic diversity studies using molecular markers except a few studies which were restricted only to rare species of ticks (Yang et al., 2004, Len et al.1996 ; Hernandez et al.,1998) in the present study, the genetic variation among seven isolated of the different species of ticks within various location of Akola district, Maharashtra in India was documented i.e. three random primers was used to analyze the close relationship.
In this study described here, we could successfully use the Pz and OPJ1 primer fragments for the 7 Indian hard tick species viz; Hyalomma a .anatolicum, Hyalomma marginatum issaci, Hyaloma hussaini, Rh. Boophilus microplus Rh. Annulatus. Rh. Haemaphysaloides, Haemaphysalis bispinosa obtained. Based on the reproducible patterns recorded for 7 species of ticks, the primers were recorded to be specific for the species. Thus, the seven studied species are having the particular genetic makeup comparable with those found elsewhere in the world. Thus the molecular taxonomy may be better than only morphological identification especially for differentiation of closely related tick species, as mentioned by Asadolla et al., (2017).
According to Mohammed and Ensan (2018) though, more comprehensive studies are needed, the primers should be valuable candidates for detection of the ticks. Our study though at preliminary level, suggest the prospective of RAPD-PCR for distinguishing ticks at the species level as well as in our better understanding; this is the first report of primers that can be used for the confirmation of tick species, in Study area. Additional study for searching intraspecific variations in specific geographical populations of the ticks from the study area will be tried in future.
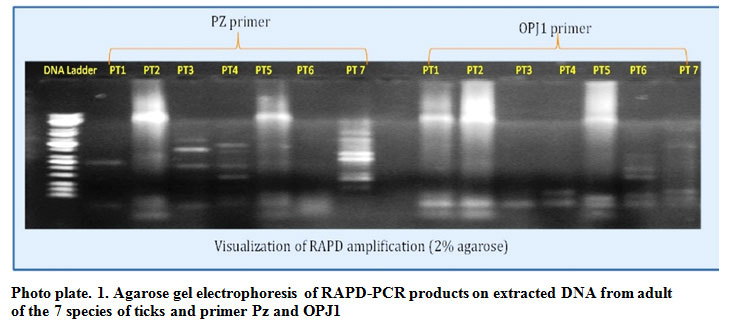 |
Plate 1: Agarose gel electrophoresis of RAPD-PCR products on extracted DNA from adult of the 7 species of ticks and primer Pz and OPJ1 |
PT1: Rh. Boophilus microplus
PT2: Hyalomma marginatum isaaci
PT3: Hyalomma a. anatolicum
PT4: Hyalomma hussaini
 |
Figure 1 |
PT5: Rh. Annulatus
PT6: Rh. Haemaphysaloides
PT7:Haemaphysalis bispinos
DNA Ladder with 100 to 10000 base pair
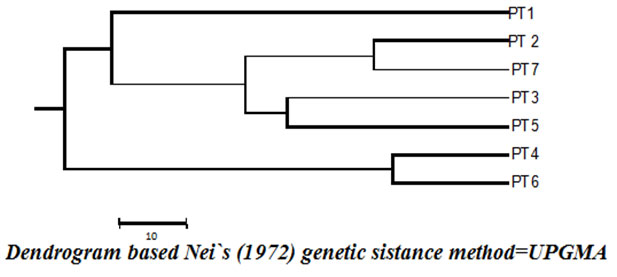 |
Figure 2 |
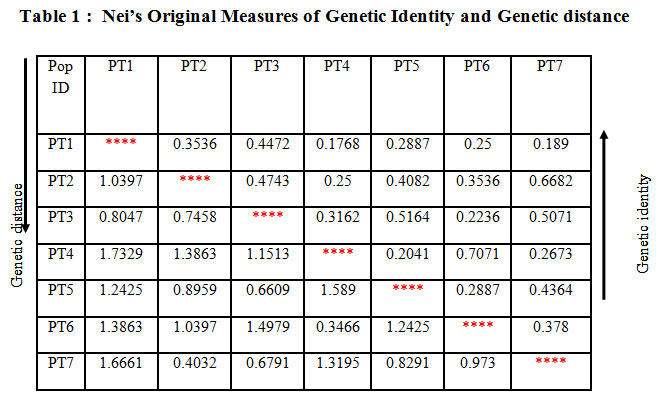 |
Table 1: Nei’s Original Measures of Genetic Identity and Genetic distance |
REFERENCES
Abdul Anvesh Mansoori, Huma Khan, Subodh Kumar Jain. (2018). RAPD-PCR based biomarker study for molecular identification and polymorphism in fish species (cyprinidae family). The Trends in Fisheries Research. Volume 7, Issue 3.
Amany M. Abd ElAzim, Etr H. K. Khashaba and Sanaa A. M. Ibrahim. (2019). Genetic polymorphism among seven entomopathogenic nematode species (Steinernematidae) revealed by RAPD and SRAP analyses. Egyptian Journal of Biological Pest Control volume 29, Article number: 17
Asadollah Hosseini-Chegeni, Mohammad Hassan Kayedi, Zakkyeh Telmadarraiy and Reza Hosseini. (2017) Multiplex-PCR differentiation of two Hyalomma and two Haemaphysalis species (Acari: Ixodidae) Persian J. Acarol., 2017, Vol. 6, No. 2, pp. 103–112
Ghosh, S., Azhahianambi P. and Yadav M. P. (2007). Upcoming and future strategies of tick control: a review. J. Vector Borne. Dis., 44: 79-89
Hernandez , R., Chen A. C, Davey R. B, Ivie G. W, Wagner G. G and George J. E (1998). Comparison of genomic DNA in various strains of Boophilus microplus (Acari: Ixodidae). J Med Entomol 35:895–900
Komalamisra, N.(1989). Genetic variability in isozymes of Anopheles minimus group from various localities from Thailand. Jpn J Sanit Zool 40:69–80
Lan, M., Sheng, W., Huang C, Gu X, Jiang Y, Zhao Y and Lu W.(1996). RAPD-PCR method for detecting DNA polymorphism in Boophilus microplus (Acari: Ixodidae). Syst Appl Acarol 1:11–14
Mohammad, Ali Al-Deeb and Mohamed Rizk Enan (2018). Genetic Diversity in the Camel Tick Hyalomma dromedarii (Acari: Ixodidae) Based on Mitochondrial Cytochrome c Oxidase Subunit I (COI) and Randomly Amplified Polymorphic DNA Polymerase Chain Reaction (RAPD-PCR). Advances in Entomology, 2018, 6, 285-294
Passos, D.T., Ferreira CAS, Da Silva S.S, Richter M. Fand Ozaki LS.(1999). Detection of genomic variability in different populations of the cattle tick Boophilus microplus in southern Brazil. Vet Parasitol 87:83–92
Stiller, D. (1992). Biotechnology: a new approach to the diagnosis and control of tick-borne hemoparasitic diseases. Ann NY Acad Sci 653:19–25
Welsh, J Mc Celland, M. (1990). Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res18:7213–7218
Williams, J. G. K., Kubelik A. R., Livak K. J., Rafalski J. A and Tingey S. V.(1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Yang, Y., Zhao, H., Di Wu, J., Zhang, J and Shi, Z. (2004) Random amplified polymorphic DNA analysis of the genomes among 7 species of ticks Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 22:223–226
Young, A. S and Morzaria, S. P. (1986) Biology of Babesia. Parasitol Today 2:211–221


