1Department of Post Graduate Studies and Research in Microbiology, Gulbarga University,
Kalaburagi-585106, Karnataka, India
2Department of Microbiology, Maharani’s Science College for Women, Bengaluru – 560 001 Karnataka, India
Corresponding author Email: smgaddad@gmail.com
Article Publishing History
Received: 12/07/2018
Accepted After Revision: 03/08/2018
Alkaline proteases are an important class of enzymes with potential industrial and commercial applications. In the present study, 52 bacterial isolates from various soil samples have been evaluated for the production of extracellular protease and selected one potent isolate based on maximum casein hydrolysis. Further, it is identified as Bacillus cereus GVK21 based on biochemical characteristics and 16S rRNA gene sequence analysis (KY659318). This organism produced 136 U/mL of protease within 48 hrs of incubation. Maximum production of protease was recorded at pH 9 and a temperature of 40°C. The present study is attempted to exploit new economical media, based on agro wastes recipes for the increased production of alkaline proteases. B.cereus GVK21 produced high levels of protease (1762 U/ml) on groundnut oil cake (1.5 %) as a substrate.The results of the study show that this isolate can be further exploited for commercial production of protease.
Protease; Bacillus Cereus; 16s Rrna; Oil Cakes; Industrial Production
M. Keshavamurthy, M. Vishwanatha, Kumar M S, Gaddad S. M. Enhanced Production of Alkaline Protease from Novel Bacterium Bacillus Cereus GVK21 Under Submerged Fermentation. Biosc.Biotech.Res.Comm. 2018;11(3).
M. Keshavamurthy, M. Vishwanatha, Kumar M S, Gaddad S. M. Enhanced Production of Alkaline Protease from Novel Bacterium Bacillus Cereus GVK21 Under Submerged Fermentation. Biosc.Biotech.Res.Comm. 2018;11(3). Available from: https://bit.ly/2KQjENK
Introduction
The emergence of new innovations are opening up new avenues in the areas of industrial biotechnology for the production of various bulk chemicals and value added products using inexpensive substrates (Binod et al., 2013). In the recent years, enzymes are replacing chemical catalysts in food, leather goods, pharmaceuticals and textiles industry(Singh et al., 2016). Proteases are a group of enzymes with wide range of applications, and account for 40 – 60% of the total enzyme sales with two thirds of them produced majorly from microorganisms (Kumar et al., 2011; Deshmukh and Vidhale, 2015).
Proteases are highly complex group of hydrolytic enzymes and occupy a pivotal status with regard to their medicinal and industrial applications (El-Bakry et al., 2015). Proteases are produced by a wide range of sources(Sharma et al., 2017). The majority of commercially available enzymes are obtained from microbial origin (Raj et al., 2012). Microbial proteases play a vital role in biotechnological processes and constitute one of the most important groups of industrial enzymes, selling product segment in the global market accounting for 60% market share (Kumar et al., 2014).Bacillus genus is one of the most important producers of extracellular proteases and industrial sectors very often use Bacillus species for the production of proteases(Contesini et al. 2018).
In the recent years, bulk chemicals and value-added products such as organic acids, amino acids, enzymes, ethanol, and single cell protein etc., are produced by the utilization of agro-industrial residues as raw materials (Pandey et al., 2000; Singhania et al., 2008). Oil cakes or oil meals are the residues obtained after oil extraction from the seeds and are rich in proteins, fiber and energy contents (Ramachandran et al.,2007).The increase in the demand leads the attention of researchers to explore novel sources for proteases, where isolation, screening and characterization of new promising strains are a continuous process. Therefore, the present study focuses on the selection of high yielding stable proteolytic bacteria from mining soil sample which is considered as an extreme environment region.
Materials And Methods
Sample Collection And Isolation
Soil samples were collected in sterile containers from different locations within the mining area of Raichur district (16o 11’ 45” North Latitude and 76o 38’ 31” East Longitude), Karnataka, India. One gram of soil was added into100 mL of enrichment medium [g/L: Casein – 10.0 and NaCl – 5.0 at pH 9.0] in a 250 mL of Erlenmeyer flask and incubated at 37o C in a rotary shaker incubator at 140 rpm for 24 hours. A loopful of the enriched medium was streaked on Nutrient agar (NA) plates and incubated at 37o C for 24 hours. Well isolated colonies were re-streaked on NA for confirming the purity and transferred onto NA slants and preserved for further use.
Screening Of The Isolates For Extracellular Protease Production
The isolates were screened for protease production on screening medium, skimmed milk agar [g/L: skim milk – 10.0; peptone – 5.0; yeast extract – 3.0; NaCl – 5.0; agar, 20 and pH 9.0](Kumar et al., 2011). After 24 hrs growth, the plates were observed for clear zones around the colonies. Proteolytic activity was further confirmed by using gelatin as the substrate in the medium (Pant et al.,2015). Depending on the zone of clearance, five isolates GVK8, GVK21, GVK29, GVK38 and GVK40, were selected for further experimental studies.
PROTEASE PRODUCTION UNDER SUBMERGED FERMENTATION
Protease production by the selected isolates was carried out by submerged fermentation. One mL of fresh bacterial inoculum was added in to 250 mL Erlenmeyer flask containing 100 mL of production medium [g/L: casein- 5.0, yeast extract – 5.0; NaCl – 5.0; MgSO4.7H2O – 0.02 and KH2PO4 – 0.05 at pH 9.0]. The flasks were placed in a rotary shaker incubator at 140 rpm at 37° C for 3-4 days. An aliquot of the culture supernatant was collected at regular time intervals of 12 hrs and assayedfor protease activity (Josephine et al.,2012). Based on the maximum protease production and shorter time required, the isolate GVK 21 is a potent strain and selected for further studies
Identification Of The Potent Protease Producing Isolate
Morphological and biochemical characteristics of the selected isolate (GVK21) was studied and recorded as per Bergey’s Manual of Systematic Bacteriology (Holt et al., 2000). The bacterial isolate was further identified by 16S rDNA sequence analysis using universal primers and genomic DNA as template. The genomic DNA of the isolate was extracted as described by Roohi et al., (2012). The PCR amplified product was sequenced at Microbial Ecology Laboratory, National Centre for Cell Science, Pune. Phylogenetic tree was constructed with Molecular Evolutionary Genetics Analysis (MEGA) 6.0 software using neighbor-joining method (Tamura et al.,2011). Duly annotated partial nucleotide sequences of the novel bacterial strain was deposited with NCBI Genbank (http://www.ncbi.nlm.nih.gov).
Scanning Electron Microscopy
Scanning Electron Microscopic (SEM) analysis was done to observe the morphology of the isolated strain.Thin film of the sample was prepared on a carbon coated copper grid by just dropping a small amount of the bacterial culture on the grid. Extra solution was removed using a blotting paper. Then the film on the SEM grid was allowed to air dry by putting it under a mercury lamp for 5 min. The sample was then observed under scanning electron microscopy TESCAN (Vega 3 LMU) at a resolution of 3 nm at various magnifications at acceleration voltage of 20.0 KV.
Protease Production In Relation To The Growth Of Bacteria
In order to study the time course for microbial growth and protease production, the isolate B. cereus GVK 21 was inoculated in the production medium and incubated in rotary shaker incubator at 160 rpm at 40° C upto 72 hours. The growth of the bacterium was determined by turbidometry, OD at 600 nm. After the removal of cells by centrifugation, the cell free supernatant was considered as the crude enzyme solution and protease activity was measured.
Evaluation Of Agro Residues (Oil Cakes) For Enhanced Protease Production
Major regional Oil cakes such as neem and pongamia were collected from local market, Bengaluru, Indiaand were fine powdered using mixer grinderand evaluated as substrates for the production of protease. Different type of oil cakes such as Castor oil cake (COC), Groundnut oil cake (GOC), Neem oil cake (NOC) and Pongamia oil cake (POC) were supplemented individually in to 100 mL of the optimized production mediumin 250 mL of Erlenmeyer flask. The agro wastes were used at a concentration of 0.5 to 2% with an increment of 0.25 %. The amount of protease produced was determined at every 6 h up to 72 h.
Protease Assay
The culture was centrifuged at 10,000 rpm for 5 min at 4° C to obtain the CFS. The protease activity of the crude enzyme was determined by the modified method of Joo et al.,(2002) briefly; 0.5 mL of CFS was added to 0.5 mL of 1% casein (as a substrate) in 0.1 M Tris-HCl (pH 9.0) and incubated at room temperature for 10 min. The reaction was terminated by the addition of 3 mL of 10% (w/v) trichloroacetic acid (TCA). The solution was centrifuged at 5000 rpm for 10 min. To the 3 mL of the clear supernatant, 5 mL 0.4 M sodium bicarbonate solution and 0.5 mL of Folin Ciocalteau reagent (FCR) were added, mixed thoroughly and incubated for 30 min at room temperature, in dark. The optical density was measured using a UV-VIS spectrophotometer (ELICO SL-159) at 660 nm against the enzyme blank. The amount of the released aromatic amino acids was calculated usingtyrosine standard.
One unit of protease is defined as the amount of the enzyme required to release 1μg of tyrosine per mL per min under the above assay conditions. Enzyme activity was calculated according to the formula of Pant et al., (2015).
Results And Discussion
Isolation, Identification, Screening And Characterization Of Protease-Producing Bacteria
In the present study, 52 independent bacteria were isolated from the soil samples collected from gold mines and were screened for proteolytic activity on skim milk agar plates (Figure 1). A total of 28 (53.84 %) isolates showed proteolytic activity ranging from 11 – 37 mm of clear zones aroundthe colonies. Five bacterial isolates, GVK8, GVK21, GVK29, GVK38 and GVK40, were selected based on the higher zone of hydrolysis on skim milk agar plates for further study. The use of skim milk agar medium for the efficient screening of proteolytic bacteria has been reported by earlier researchers (Raj et al., 2012; Ravi et al., 2015).
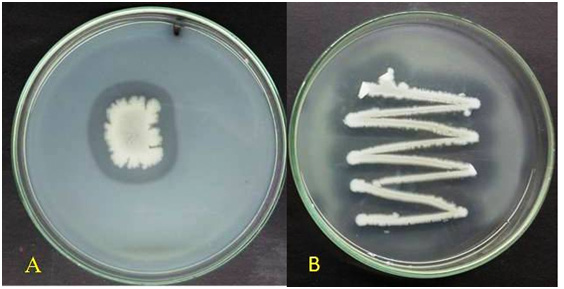 |
Figure 1: Zone of hydrolysis on A) skim milk agar B) Gelatin agar by bacterial isolate Bacillus cereus GVK 21. |
The selected five isolates were further screened for the quantitative production of the enzyme in production medium (Figure 2). The protease production was slower during the first 12 hours by all the isolates. The protease production increased significantly in respect of the isolate GVK21 during 12 to 24 hours to reach the maximum activity and remained at that level till 48 hours. While with respect to the other isolates, the protease activity increased gradually and the maximum activities were recorded at 48 hours of incubation. This indicated that the isolate GVK21 produces a maximum protease activity of 136 U/mL and takes shorter time in preliminary study.
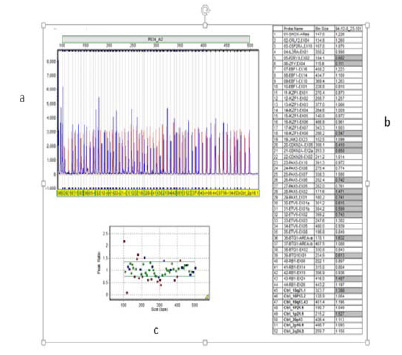 |
Figure 2: Quantitative production of protease using different bacterial isolates. |
The strain GVK21 is gram positive rod, motile, spore former and was tentatively identified as Bacillus sp. based on its morphological and biochemical characteristics (Table 1), (Holt et al.,2000). The 16S rDNA was amplified through PCR which showed1500 kb band on 2% agarose gel (Figure 3). Subsequently, the comparison of the 16S rRNA gene nucleotide sequence (1387 bp) of the strain GVK21 with other 16S rRNA genes sequences of closely related strains from NCBI database showed that this isolate has 99 % sequence homology with B. cereus ATCC 14579(Accession No. 074540). The phylogenetic tree, constructed by the neighbor-joining method indicated that the strain GVK21 is affiliated with the genus Bacillus and closely related to B. cereus strain LG1 – Accession No. KF307764 (Figure 4). The obtained nucleotide sequence GVK21 was submitted to GenBank database and the accession number assigned is KY659318 (https://www.ncbi.nlm.nih.gov/ nuccore/KY659318). The species B. cereusATCC 14579(074540.1), B. cereus strain JCM 2152 (113266.1), B. thuringiensis strain NBRC 101235 (112780.1) has the closest sequence similarity of 99%.
| Table 1: Morphological and biochemical characteristics of B. cereus strain GVK21 isolated from soil. |
|
| Morphological Characteristics | Results |
| Gram staining | Positive rods |
| Colour | Creamish white |
| Motility test | Motile |
| Spore | Spore former |
| Physiological characteristics | |
| Catalase | Positive |
| Indole | Negative |
| Methyl red | Negative |
| Voges Proskauer | Positive |
| Citrate utilization | Positive |
| Oxidase reaction | Positive |
| Casein hydrolysis | Positive |
| Gelatin liquefaction | Positive |
| Starch hydrolysis | Positive |
| Nitrate reduction | Positive |
| Growth at 4o C | – |
| Growth at 40o C | + |
 |
Figure 3: Agarose Gel Electrophoresis of B.cereus DNA. Lane1- 100 bp DNA ladder, Lane2- 16S rDNA amplicon of B.cereus GVK21 |
 |
Figure 4: Phylogram obtained based on phylogenetic analysis of 16S rDNA gene sequence data showing the phylogenetic positions of isolate Bacillus cereus strain GVK21 and of a number of related taxa |
Bacillus is an industrially important organism and was the first to be used in the commercial production of protease in 1952 (Binod et al., 2013). Several proteases have been produced from many Bacillus species (Rao et. al., 1998). Bacillus species like B. subtilis (Babe and Schmidt, 1998), B. licheniformis (Mabrouk et al., 1999), B. sphaericus (Singh et al., 1999), B. proteolyticus (Bhaskaret al., 2007), Bacillus cereus (Doddapaneni et al., 2009; Bajaj et al., 2013) and B. megaterium (Rajkumaret al., 2010) have been reported for protease production. Other than Bacillus species bacteria such as Pseudomonas fluorescens (Kalaiarasi and Sunitha, 2009), P. aeruginosa (Raj et al., 2012), Vibrio etschnikovii (Jellouliet al., 2009), V. alginolyticus (Shanthakumari et al., 2010) are also reported for protease production.
Bacillus cereus GVK 21 was subjected for secondary screening for quantitative protease production under submerged condition. The protease production increased significantly from 12 U/mL to 136 U/mL during6 h to 36 h of incubation and the enzyme activity was in synchrony with the growth of the bacterium, wherein the logarithmic growth was observed during 6 to 24 hours and the maximum growth was observed at 36 hours indicating that the protease production is maximum in the late exponential and early stationary phase (Figure 5). The decline in the protease production was observed after 48 h. Kannikar et al.,(2008) reported that Bacillus sp. BA40 produced 1.158 U/mL protease activity, B. licheniformis LBBL-11 showed 18.4 U/mL at 48 hours (Olajuyigbe, and Ajele,2008). The results indicate that the isolate B. cereus GVK 21 is producing 136U/mL a comparatively higher protease activity, that too under preliminary screening.
 |
Figure 5: Protease production with relation to growth of Bacillus cereus GVK 21. |
Chitte and Dey (2000) have also shown that the log phase the optimum for the production ofprotease by Streptomyces megasporus. Incubation time of 24 hours positively influences the production of protease from Bacillus cereus SRM-001 (Narasimhan et al.,2015). Incubation time of 40 h was found to enhance protease production by Bacillus natto-NRRL-3666 (Mahajan et al.,2010).
Sem Analysis
Figure 6 shows SEM image of bacteria which confirmed the rod shaped nature of the cells, where the cell size (length and breadth) is found to be 1.91 µm and 0.82 µm respectively.
 |
Figure 6: SEM images of B.cereus GVK 21 |
Enhanced Protease Production Using Oil Cakes
The effect of agro-based by products as alternative substrates for bacterial protease production under submerged fermentation has been studied by several workers (Praveen Kumar et al., 2008, Prasad, et al., 2014). B. cereus GVK21 (KY659318) produced varying levels of alkaline protease on various agriculture based substrates. Among various oil cakes (Figure. 7) examined, groundnut oil cake enhanced the protease production from B. cereus GVK21 by 112 % (1762 U/ml) as compared to control (665 U/ml), i.e., groundnut oil cake supported maximum protease production. Bacillus subtilis SHS-04exhibited maximum protease production (1616.21 U/mL) by utilizing groundnut cake as substrate (Olajuyigbe, 2013).
 |
Figure 7: Different Oil cakes used as a substrate for protease production. |
Other oil cakes also substantially supported protease production, castor cake (1316 U/ml), pongamia cake (1144 U/ ml) and neem cake (904 U/ ml) as shown in Figure 8. Enzyme synthesis was found to be repressed by rapidly metabolizable nitrogen sources such as amino acids or ammonium ion concentrations in the medium as also observed in the presence of ammonium sulphate and potassium nitrate (Saurabh et al., 2007; Bajaj and Sharma, 2011). Therefore complex nitrogen sources are usually used for protease production. Groundnut oil cake, a rich protein source (30–40%), which mainly constitutes different amino acids such as arginine (11.0), Leucine (6.1), glycine (6.0), and phenyl alanine (4.9) and a dry matter of 92.6% comprehends slow release of nitrogen due to its conditioned and moderately complex nature which may favor the process organism metabolically and physiologically for efficient production of protease (Kuo, 1967). Several reports indicate that groundnut cake serves as a reasonably good nitrogen source for protease production by various microorganisms (Kuberan et al., 2010; Kranthi et al., 2013, Olajuyigbe, 2013).
 |
Figure 8: Enhanced Protease production using different oil cakes |
The substrate in the growth medium constitutes a major cost determining factor for the commercial production of industrial enzymes. The high cost of protease production is another major hindrance for wide range of industrial and medicinal applications of this enzyme (Jayasree et al., 2009). Utilization of agro industrial residues as carbon and nitrogen sources for the bulk production of industrial enzymes may play a significant role not only in reducing the production cost and also contribute towards growing environmental concerns by addressing the agro industrial waste disposal and management problems because of tremendous quantities of agricultural residues generated through agricultural practices and industrial processes (Bajaj and Wani, 2011; Singh and Bajaj 2015). Earlier workers also reported that defatted oil meals like soybean meal (Saurabh et al., 2007), cotton seed cake (Bajaj et al., 2013) and groundnut meals (Olajuyigbe, 2013) are the cost-effective alternatives for industrial production processes. Hence utilization of a low-priced nitrogen source is an important criterion for economic production of industrial enzymes. Requirement of specific nitrogen source differs from one organism to other even among the same species isolated from different sources (Bajaj and Sharma, 2011).
Conclusion
Bacillus cereus GVK 21 (KY659318) isolated from the soil samples from a mining area exhibited higher proteolytic activity of 136 U/mL which significantly increased to 1762 U/mL on ground nut cake, comparatively higher than already reported and thus can be exploited as a potential source for large scale production of protease enzyme to cope up the needs of industrial applications and the demand of the global market.
Acknowledgments
Authors wish to acknowledge Dr. Yogesh Souche, Scientist, National Centre for Cell Science, Govt of India, Pune for sequencing and identification of Bacteria and B.M.S College of Engineering, Bengaluru, Karnataka, India for SEM analysis. Authors are thankful to the Maharani’s Science College for Women, Bengaluru, Karnataka, India for providing facilities to carry out this work.
Conflict of Interest
Authors declare that they have no conflict of interest in the publication.
References
Babe, L., and Schmidt, B., 1998. Purification and biochemical analysis of WprA, a 52-kDa serine protease secreted by B. subtilis as an active complex with its 23-kDa propeptide. Biochimicaet Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology, 1386(1),pp.211-219.
Bajaj, B.K. and Sharma, P., 2011. An alkali-thermotolerant extracellular protease from a newly isolated Streptomyces sp. DP2. New biotechnology, 28(6), pp.725-732.
Bajaj, B.K. and Wani, M.A., 2011. Enhanced phytase production from Nocardia sp. MB 36 using agro‐residues as substrates: Potential application for animal feed production. Engineering in Life Sciences, 11(6), pp.620-628.
Bajaj, B.K., Sharma, N. and Singh, S., 2013. Enhanced production of fibrinolytic protease from Bacillus cereus NS-2 using cotton seed cake as nitrogen source. Biocatalysis and Agricultural Biotechnology, 2(3), pp.204-209.
Bhaskar, N., Sudeepa, E.S., Rashmi, H.N. and Selvi, A.T., 2007. Partial purification and characterization of protease of Bacillus proteolyticus CFR3001 isolated from fish processing waste and its antibacterial activities. Bioresource Technology, 98(14), pp.2758-2764.
Binod, P., Palkhiwala, P., Gaikaiwari, R., Nampoothiri, K.M., Duggal, A., Dey, K. and Pandey, A., 2013. Industrial Enzymes-Present status and future perspectives for India.
Chitte, R.R. and Dey, S., 2000. Potent fibrinolytic enzyme from a thermophilic Streptomyces megasporusstrain SD5. Letters in applied microbiology, 31(6), pp.405-410.
Contesini, F.J., Melo, R.R.D. and Sato, H.H., 2018. An overview of Bacillus proteases: from production to application. Critical reviews in biotechnology, 38(3), pp.321-334.
Doddapaneni, K.K., Tatineni, R., Vellanki, R.N., Rachcha, S., Anabrolu, N., Narakuti, V. and Mangamoori, L.N., 2009. Purification and characterization of a solvent and detergent-stable novel protease from Bacillus cereus. Microbiological Research, 164(4), pp.383-390.
El-Bakry, M., Abraham, J., Cerda, A., Barrena, R., Ponsá, S., Gea, T. and Sánchez, A., 2015. From wastes to high value added products: novel aspects of SSF in the production of enzymes. Critical Reviews in Environmental Science and Technology, 45(18), pp.1999-2042.
Holt, J.G., Krieg, N.R., Sneath, P., Staley, J.T. and Williams, S.T., Bergey’s Manual of Determinative Bacteriology. 2000. Lippincot Williams & Wilkins. pag, pp.571-572.
Jayasree, D., Kumari, T.D.S., Kishor, P.B.K., Lakshmi, M.V. and Narasu, M.L., 2009. Optimization of production protocol of alkaline protease by. Streptomyces pulvereceus; Inter JRI Science and Technology, 1(2).
Jellouli, K., Bougatef, A., Manni, L., Agrebi, R., Siala, R., Younes, I. and Nasri, M., 2009. Molecular and biochemical characterization of an extracellular serine-protease from Vibrio metschnikovii J1. Journal of industrial microbiology & biotechnology, 36(7), pp.939-948.
Joo, H.S., Kumar, C.G., Park, G.C., Kim, K.T., Paik, S.R. and Chang, C.S., 2002. Optimization of the production of an extracellular alkaline protease from Bacillus horikoshii. Process Biochemistry, 38(2), pp.155-159.
Josephine, F.S., Ramya, V.S., Devi, N., Ganapa, S.B. and Vishwanatha, T., 2017. Isolation, production and characterization of protease from Bacillus sp isolated from soil sample. Journal of Microbiology and Biotechnology Research, 2(1), pp.163-168.
Kalaiarasi, K. and Sunitha, P.U., 2009. Optimization of alkaline protease production from Pseudomonas fluorescens isolated from meat waste contaminated soil. African Journal of Biotechnology, 8(24).
Kranthi, V.S., Rao, D.M. and Jaganmohan, P., 2012. Protease production by Rhizopus stolonifer through solid state fermentation. Cent. Euro. J. Exp. Bio, 1, pp.113-117.
Kumar, E.V., Srijana, M., Kumar, K.K., Harikrishna, N. and Reddy, G., 2011. A novel serine alkaline protease from Bacillus altitudinis GVC11 and its application as a dehairing agent. Bioprocess and Biosystems engineering, 34(4), pp.403-409.
Kumar, D., Kumar, V., Verma, A.K. and Dubey, A., 2013. Kinetic characterization and immobilization of partially purified extracellular alkaline protease from rhizospheric soil bacterium Bacillus subtilis strain EN4. J. Pure Appl. Microbiol, 7(1), pp.727-732.
Kumar, P.P., Mathivanan, V., Karunakaran, M., Renganathan, S. and Sreenivasan, R.S., 2008. Studies on the effects of pH and incubation period on protease production by Bacillus spp. using groundnut cake and wheat bran. Indian Journal of Science and Technology, 1(4), pp.1-4.
Mabrouk, S.S., Hashem, A.M., El-Shayeb, N.M.A., Ismail, A.M. and Abdel-Fattah, A.F., 1999. Optimization of alkaline protease productivity by Bacillus licheniformis ATCC 21415. Bioresource Technology, 69(2), pp.155-159.
Maghsoodi, V., Kazemi, A., Nahid, P., Yaghmaei, S. and Sabzevari, M.A., 2013. Alkaline protease production by immobilized cells using B. licheniformis. Scientia Iranica, 20(3), pp.607-610.
Mahajan, P.M., Gokhale, S.V. and Lele, S.S., 2010. Production of nattokinase using Bacillus natto NRRL 3666: media optimization, scale up, and kinetic modeling. Food Science and Biotechnology, 19(6), pp.1593-1603.
Narasimhan, M.K., Chandrasekaran, M. and Rajesh, M., 2015. Fibrinolytic enzyme production by newly isolated Bacillus cereus SRM-001 with enhanced in-vitro blood clot lysis potential. The Journal of general and applied microbiology, 61(5), pp.157-164.
Olajuyigbe, F.M. and Ajele, J.O., 2008. Some properties of extracellular protease from Bacillus licheniformis LBBL-11 isolated from iru, a traditionally fermented African locust bean condiment. African Journal of Biochemistry Research, 2(10), pp.206-210.
Olajuyigbe, F.M., 2013. Optimized production and properties of thermostable alkaline protease from Bacillus subtilis SHS-04 grown on groundnut (Arachis hypogaea) meal. Advances in Enzyme Research, 1(04), p.112.
Pandey, A., Soccol, C.R., Nigam, P., Soccol, V.T., Vandenberghe, L.P. and Mohan, R., 2000. Biotechnological potential of agro-industrial residues. II: cassava bagasse. Bioresource technology, 74(1), pp.81-87.
Pant, G., Prakash, A., Pavani, J.V.P., Bera, S., Deviram, G.V.N.S., Kumar, A., Panchpuri, M. and Prasuna, R.G., 2015. Production, optimization and partial purification of protease from Bacillus subtilis. Journal of Taibah University for Science, 9(1), pp.50-55.
Prasad, R., Abraham, T.K. and Nair, A.J., 2014. Scale up of production in a bioreactor of a halotolerant protease from moderately halophilic Bacillus sp. isolated from soil. Brazilian Archives of Biology and Technology, 57(3), pp.448-455.
Raj, A., Khess, N., Pujari, N., Bhattacharya, S., Das, A. and Rajan, S.S., 2012. Enhancement of protease production by Pseudomonas aeruginosa isolated from dairy effluent sludge and determination of its fibrinolytic potential. Asian Pacific Journal of Tropical Biomedicine, 2(3), pp.S1845-S1851.
Rajkumar, R., Jayappriyan, K.R., Kannan, P.R. and Rengasamy, R., 2010. Optimization of Culture Conditions for the Production of Protease from Bacillus megaterium. Journal of Ecobiotechnology.
Rao, M.B., Tanksale, A.M., Ghatge, M.S. and Deshpande, V.V., 1998. Molecular and biotechnological aspects of microbial proteases. Microbiology and molecular biology reviews, 62(3), pp.597-635.
Ramachandran, S., Singh, S.K., Larroche, C., Soccol, C.R. and Pandey, A., 2007. Oil cakes and their biotechnological applications–A review. Bioresource technology, 98(10), pp.2000-2009.
Ravi, M., Rayudu, K., Gaddad, S.M. and Jayaraj, Y.M., 2015. Studies on the potent protease producing bacteria from soil samples. Int. J. Curr. Microbiol. App. Sci, 4(1), pp.983-988.
Rupali R. Deshmukh and Vidhale N N.,2015. Effect of pH on the production of protease by Fusarium oxysporum using agroindustrial waste. Biosci. Biotech. Res. Comm. 8(1): 78-83 (2015)
Santong, K., Chunglok, W., Lertcanawanichakul, M. and Bangrak, P., 2011. Screening and isolation of Bacillus sp. producing thermotolerant protease from raw milk. Walailak Journal of Science and Technology (WJST), 5(2), pp.151-160.
Saurabh, S., Jasmine, I., Pritesh, G. and Kumar, S.R., 2007. Enhanced productivity of serine alkaline protease by Bacillus sp. using soybean as substrate. Malaysian Journal of Microbiology, 3(1), pp.1-6.
Shanthakumari, A.R., Nagalakshmi, R. and Ramesh, S., 2010. Scaleup and media optimization of protease by Vibrio alginolyticus. Journal of Ecobiotechnology.
Sharma, K.M., Kumar, R., Panwar, S. and Kumar, A., 2017. Microbial alkaline proteases: Optimization of production parameters and their properties. Journal of Genetic Engineering and Biotechnology, 15(1), pp.115-126.
Singh, J., Vohra, R.M. and Sahoo, D.K., 1999. Alkaline protease from a new obligate alkalophilic isolate of Bacillus sphaericus. Biotechnology letters, 21(10), pp.921-924.
Singh, S. and Bajaj, B.K., 2015. Medium optimization for enhanced production of protease with industrially desirable attributes from Bacillus subtilis K-1. Chemical Engineering Communications, 202(8), pp.1051-1060.
Singh, R., Kumar, M., Mittal, A. and Mehta, P.K., 2016. Microbial enzymes: industrial progress in 21st century. 3 Biotech, 6(2), p.174.
Singhania, R.R., Soccol, C.R. and Pandey, A., 2008. Application of tropical agro-industrial residues as substrate for solid-state fermentation processes. In Current developments in solid-state fermentation (pp. 412-442). Springer, New York, NY.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S., 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution, 28(10), pp.2731-2739.


