1Department of Biotechnology, Hemchandracharya North Gujarat University, Patan, Gujarat, India
2Dr.Indu Dayal Meshri College of Science and Technology, Patan, Gujarat, India
Corresponding author email: arpit3290@gmail.com
Article Publishing History
Received: 19/10/2019
Accepted After Revision: 18/12/2019
Dengue fever is caused by Dengue Virus (DENV) and it has become a serious threat to human lives. This study was aimed at in-silico drug design and prediction of Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties of potential hits against dengue fever using nonstructural protein 3(NS3). NS3 plays a vital role in the life cycle of dengue virus. It is a target for the development of specific antiviral inhibitors. The 3D structure of Dengue Virus 4 NS3 Helicase in complex with ssRNA is available at Protein Data Bank. It contains Glycerol as a ligand. Compounds were selected from DrugBank, Pub Chem, Chem Spider and they were screened along with Glycerol using PyRx-Virtual Screening software for Structure based drug design (SBDD) and compared using similarity score. Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties were determined by using online tool Danish Quantitative Structure-Activity Relationship (QSAR) database. The Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) analysis and docking results revealed 12 compounds as potential leads.
ADMET, Dengue Virus 4, NS3 Helicase – ssRNA complex.
Trivedi A, Dave S. M, Patel J. Design and Prediction of Absorption, Distribution, Metabolism, Excretion and Toxicity Properties of Drugs for Dengue fever Using in-silico Approaches. Biosc.Biotech.Res.Comm. 2019;12(4).
Trivedi A, Dave S. M, Patel J. Design and Prediction of Absorption, Distribution, Metabolism, Excretion and Toxicity Properties of Drugs for Dengue fever Using in-silico Approaches. Biosc.Biotech.Res.Comm. 2019;12(4). Available from: https://bit.ly/379u9nk
Copyright © Trivedi et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Severe dengue fever (otherwise called Dengue Haemorrhagic Fever) was first perceived during the 1950s during dengue epidemics in the Philippines and Thailand. Today, it affects most Asian and Latin American nations and has turned into a main leading cause of hospitalization and death among children and adults in these regions(Bollati et al., 2010; Qaddir, Rasool, Hussain, & Mahmood, 2017).
As indicated by World Health Organization (WHO), dengue is a mosquito borne viral infection that has quickly spread in all locales as of late. This virus is transmitted by female mosquitoes mainly of the species Aedes aegypti and, to a lesser degree, Aedes albopictus(“Dengue Bulletin,” 2016). This mosquito additionally transmits chikungunya, yellow fever and zika disease. Dengue is boundless all through the tropics, with local variation in risk affected by rainfall, temperature and impromptu rapid urbanization(“Dengue Bulletin,” 2016).
There are 4 distinct, however firmly related, serotypes of the virus that cause dengue (DEN-1, DEN-2, DEN-3 and DEN-4)(Mukhopadhyay, Kuhn, & Rossmann, 2005; Wilschut, 2010).Recuperation from disease by one gives lifelong immunity against that specific serotype. Be that as it may, cross-immunity to the other serotypes after recovery is only partial and temporary. Subsequent infections by other serotypes increase the risk of developing severe dengue(Kuhn et al., 2002). It is an acute infectious disease characterized by headache, pain in various parts of the body, fever, rash, leucopenia and so on(Alvarez, Laura, Ezcurra, Fucito, & Gamarnik, 2005).The instance of dengue fever has expanded in ongoing time in different geographic areas with the distribution of the viruses and the mosquito vectors and it has led to build pandemic activity, the development of hyper endemicity and the emergence of dengue hemorrhagic fever in new geographic regions(Whitehead, Blaney, Durbin, & Murphy, 2007).
Dengue viruses enter and replicate with the help of certain structural and nonstructural proteins in cells of mononuclear phagocyte to further increase the Dengue Virus infection in these cells(Mazzon, Jones, Davidson, Chain, & Jacobs, 2009).Dengue infection can cause fever of different severity. At the point when the distinctive strains of dengue virus connect with the immune system of different person it can cause complex interaction resulting in conditions such as dengue fever or dengue shock syndrome. At present there is not any treatment available, in the form of vaccine or drug. The control of mosquitoes has shown only limited success(Qi, Zhang, & Chi, 2008).
Therefore, there is a need to develop novel drugs for Dengue fever. One of the most promising target for development of novel drugs is the dengue non-structural protein 3 (NS3), the biggest and most highly conserved dengue proteins(Ackermann & Padmanabhan, 2001).The NS3 protein of Dengue Virus is a multifunctional protein having molecular weight of 69 kDa, provided with helicase, protease, and nucleoside 5-triphosphatase (NTPase) activities(Ago et al., 1999).The computer-aided drug design is a specialized branch that utilizes computational strategies to create drug-receptor interactions(Bissantz, Folkers, & Rognan, 2000).The computer-aided drug design methods are greatly dependent on the tools of bioinformatics, applications and databases.
Since the structure of the NS3 is available, structure based drug design approaches are best suited for novel drug design to identify potential hits and lead molecules(Bissantz et al., 2000).
Most of drug hopefuls fail in Phase III clinical trials after many years of research. The disappointment is due to toxicity or problems with metabolism. The ADMET properties of drugs are Absorption, Distribution, Metabolism, Excretion and Toxicity in other words bioavailability and bioactivity. However these properties are typically measured in the wet lab, they can likewise be normal ahead of time with the assistance of bioinformatics software and tools. The range of bioinformatics apparatuses and web indexes (search engines) are available for this work (Bissantz et al., 2000; Iskar, Zeller, Zhao, Noort, & Bork, 2011).
Once potential lead molecules have been identified the next stage is to fine tune their structural and ADMET properties. This commonly includes the arrangement of changes in accordance with the essential and secondary structure of the compound. The procedure can be improved by utilizing software that explores related compounds to the lead candidate(Song, Lim, & Chuantong, 2009).
MATERIAL AND METHODS
Target Identification:Dengue Virus contains structural and nonstructural proteins. Among all these proteins non-Structural protein named Dengue Virus 4 NS3 Helicase in complex with ssRNA (figure 1) was identified as target.
 |
Figure 1: Dengue Virus 4 NS3 Helicase in complex with ssRNA |
The structure of the protein molecule was downloaded in from Protein Data Bank (PDB) with PDB ID 2JLU. The experimental detail shows, Resolution[Å]: 2.04, R-Value: 0.197 (obs.) and R-Free:0.242.
Drug Library:Online chemical databases throughout the web were employed to obtain compounds. This mixes were screened which had related structures and properties with Glycerol, (propane-1,2,3-triol) using similarity score. Such properties like Common and IUPAC name of Compounds, Melting and Boiling Point of Compounds, pKa Value, Polarizability, Molecular Weight, Solubility, Density have been already reported(Dias, Filgueira, & Jr, 2008; Fernando et al., 2008).
Docking: Structure files such as .sdf/.mol/.pdb of those chemical compounds were downloaded from the chemical compound database like PubChem, DrugBank, ChemSpider. Docking was performed on the structure files of the compounds by using docking module PyRx. Binding energy was obtained as a result of the docking process that is shown in Table 1. These compounds were further analyzed for their Absorption, Distribution, Metabolism, Excretion and Toxicity properties.
Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET):ADMET properties of chemical compounds were examined by online database named Danish Quantitative Structure-Activity Relationship (QSAR) database. Multicase Acute Aquatic Toxicity, Carcinogenicity, Arylhydroxylase Activity, Lethal Body Burden, Bioconcentration, Mutagenicity, Biodegradation, Environmental Partitioning and General Properties are included in ADMET properties. Forty compounds have affirmed ADMET properties using software but out of these forty compounds twelve compounds lastly fulfill maximum of the characteristics of drug target hopefuls. Thus, binding energy and ADMET of those twelve compounds have been shown in the outcome tables (Table:1 and Table:2).
Table 1: Showing Binding Energy Of Chemical Compounds
| Drug Accession Number | Binding Energy
kca/mol |
| DB00825 | -5.6 |
| DB02202 | -3.7 |
| DB02659 | -8.6 |
| DB02854 | -8.6 |
| DB02901 | -7.3 |
| DB03017 | -4.5 |
| DB03193 | -4.6 |
| DB03500 | -5.6 |
| DB03619 | -8.6 |
| DB06777 | -8.3 |
| DB11789 | -8.6 |
| DB13587 | -8.0 |
Table 2: Absorption, Distribution, Metabolism, Excretion and Toxicity Properties
| Health End Point | Mutagenicity | In Vitro Tests | Carcinogenicity | |||
| Severe skin irritation
|
Skin sensitization
|
Respiratory sensitization | Ames test (Salmonella) | HGPRT | ||
| METHANOL – (DB00825) | NEG | NEG | NEG | NEG | NEG | NEG |
| 1,3BUTANEDIOL – (DB02202) | NEG | NEG | NEG | NEG | NEG | NEG |
| CHOLICACID – (DB02659) | NEG | NEG | NEG | POS | NEG | NEG |
| LAURIC ACID – (DB03017) | NEG | NEG | POS | NEG | NEG | NEG |
| STERIC ACID – (DB03193) | NEG | NEG | POS | NEG | NEG | NEG |
| Aetiocholanolone (DB02854) | NEG | NEG | POS | NEG | NEG | NEG |
| TRICOSANIC ACID – (DB03500) | NEG | NEG | NEG | POS | NEG | POS |
| DEOXYCHOLIC ACID – (DB03619) | NEG | NEG | NEG | NEG | NEG | NEG |
| Chenodeoxycholic acid – (DB06777) | NEG | NEG | NEG | NEG | NEG | NEG |
| STANOLONE – (DB02901) | NEG | NEG | NEG | NEG | NEG | NEG |
| MESTEROLONE – (DB13587) | NEG | NEG | NEG | POS | NEG | NEG |
| HYDEOXYCOLIC ACID – (DB11789) | NEG | NEG | NEG | NEG | NEG | NEG |
( * POS = POSITIVE) (* NEG =NEGATIVE)
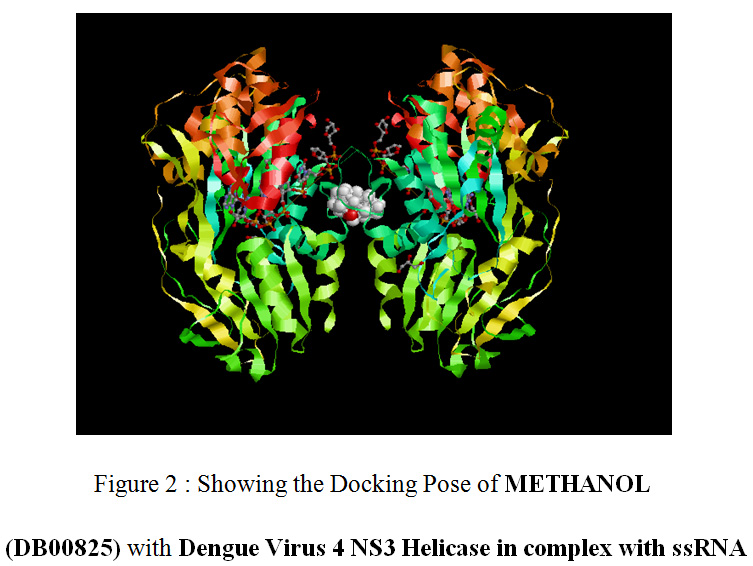 |
Figure 2: Showing the Docking Pose of METHANOL (DB00825) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
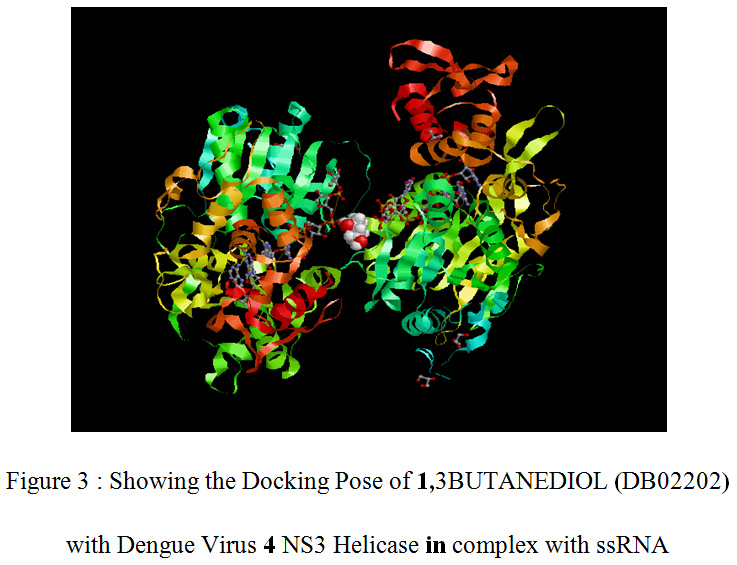 |
Figure 3 : Showing the Docking Pose of 1,3BUTANEDIOL (DB02202) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
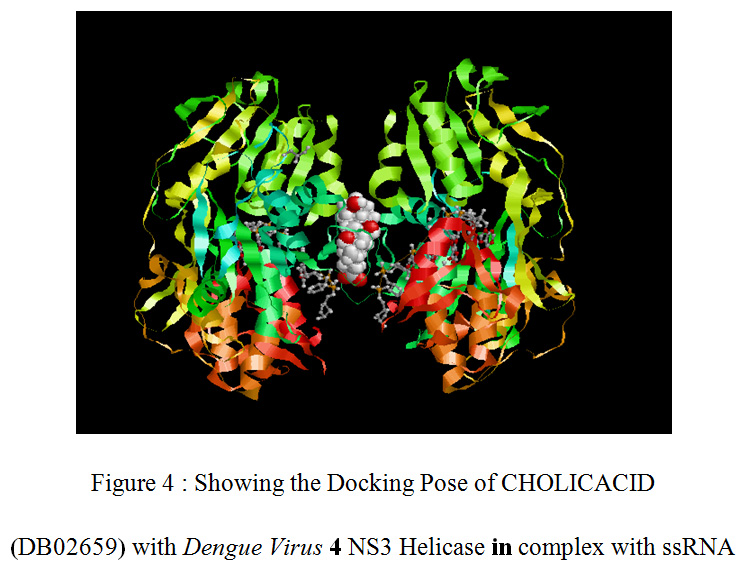 |
Figure 4 : Showing the Docking Pose of CHOLICACID (DB02659) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 5 : Showing the Docking Pose of Aetiocholanolone (DB02854) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 6 : Showing the Docking Pose of Stanolone (DB02901) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 7 : Showing the Docking Pose of LAURIC ACID (DB03017) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 8 : Showing the Docking Pose of STERIC ACID (DB03193) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 9 : Showing the Docking Pose of TRICOSANIC ACID (DB03500) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 10 : Showing the Docking Pose of DEOXYCHOLIC ACID (DB03619) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 11 : Showing the Docking Pose of Chenodeoxycholic acid (DB06777) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 12 : Showing the Docking Pose of HYDEOXYCOLIC ACID (DB11789) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
 |
Figure 13 : Showing the Docking Pose of MESTEROLONE (DB13587) with Dengue Virus 4 NS3 Helicase in complex with ssRNA |
RESULTS AND DISCUSSION
In this work total ninety chemical compounds were screened as drug target candidate for Dengue Virus protein 4 NS3. Chemical and physical properties of these compounds were collected from chemical database. These properties such as pKa value, Molecular weight(MW), its state, Polarizibility, Melting and Boiling Point play essential role in the drug molecule(Dias et al., 2008).
A weak acid incorporates a pKa worth within the scope between -2 to 12 and pKa values over twelve area unit are said to be alkaline drug. Acid with a pKa worth of less concerning -2 area units are said to be a strong acid. Similarity score denotes relevance of the new chemical with the chemical which may fit the drug target pocket.Polarizability shows the ability of a molecule to be polar; it is dependent upon the charge distribution and effects the bond formation in a chemical compound. Docking results denote that lower binding energy show the higher affinity of the drug target candidate. –5 to -9 Kcal/Mol is the normal range of binding energy(Modis, Ogata, Clements, & Harrison, 2003).
Every drug must comply with its ADMET properties. Amongst all their properties Mutagenicity, Toxicity, In-vitro test and Carcinogenicity are important properties of drug candidates to be examined. Preferably these properties are examined by tests like Ames test, Food and Drug Administration’s Center for Drug Evaluation and Research (FDA- CDER) properties on RAT and Mouse, Lethal Concentration, Lethal Dose in human, and Hypoxanthine-guannine phosphoribosyltransferase (HGPRT) test. These properties can also be predicted using ADMET software(Yang & Chen, 2004).
Table 2 enlists skin irritation, skin sensitization, Respiratory sensitization and other properties of these molecules. Most of the molecules show negative skin sensitization, skin irritation, respiratory irritation and negative Ames and Hypoxanthine-guannine phosphoribosyltransferase (HGPRT) test indicates that these molecules can be considered as suitable hits for further drug design.
Ames test can be utilized to check mutagenicity of the drug. Positive result indicates that the drug will be the mutagenic(Wadood et al., 2013; Williamson, 2014). CHOLICACID – (DB02659),MESTEROLONE – (DB13587)chemical compounds demonstrate positive Ames test, thereupon they are mutagenic and can only be used as drug target candidates after bringing changes in their molecular structure. Remaining compounds show negative Ames test; thus they are not mutagenic and can be considered as drug target candidates.
Certain tests such as-CDER Proprietary Male Rat, FDA Cancer Male Rat, FDA Cancer Female Rat, FDA Cancer Male Mouse, CDER Proprietary Male Mouse, FDA Cancer Female Mouse, -CDER Proprietary Female Rat, -CDER Proprietary Female Mouse are used to determine carcinogenicity. Positive result denotes that the drug will be the carcinogenic(Wadood et al., 2013; Williamson, 2014). TRICOSANIC ACID – (DB03500), chemical compounds indicate positive carcinogenicity thence they can only be considered as drug candidates after modification in the structure. Remaining compounds show negative carcinogenicity thus they are not carcinogenic and can be considered as drug target candidates.
Lethal dose is a way to determine the short term poisoning potential of the material. Lethal concentration values solicit the concentration of the chemical. This toxicity is established on lethal concentration of the drug. The range of lethal concentration 100 – 1000mg/L indicates moderately toxicity of compound and based on that concentration lethal dose is set as 0.5-5 gm/kg. If lethal concentration range is 10 -100 mg/L then it indicates highly toxicity of the substance and hence lethal dose has to be 5-50 mg /kg. If lethal concentration range is 1000 – 10000 mg/L then it indicates slightly toxicity of the compounds and lethal dose is set as 5 – 15 gm/ kg(Wadood et al., 2013; Williamson, 2014).
Here screened each of the twelve compounds have lethal concentration range 100 – 1000 mg/L thus they are sensibly toxic and their lethal dose should be 0.5 – 5 gm/Kg. After evaluating physical and chemical properties, docking and ADMET analysis of all the screened chemical compounds we validate below chemical compounds as potential lead molecules for Dengue Virus4 NS3 protein. METHANOL – (DB00825), 1,3BUTANEDIOL – (DB02202), CHOLICACID – (DB02659), LAURIC ACID – (DB03017), STERIC ACID – (DB03193), Aetiocholanolone– (DB02854), TRICOSANIC ACID – (DB03500), DEOXYCHOLIC ACID – (DB03619), Chenodeoxycholic acid– (DB06777), Stanolone – (DB02901), HYDEOXYCOLIC ACID – (DB11789), MESTEROLONE – (DB13587).Chemical Compounds apart from these are missing in above criteria. Thus, those chemical compounds can’t be employed in present form as drug candidate for the same target.
SUMMARY AND CONCLUSION
Dengue Virus protein replicates in the host cells. NS3 Helicase in complex with ssRNA contains ligand named Glycerol, (propane-1,2,3-triol). It was identified as a drug target protein. Its structure was downloaded from the Protein Data Bank. Ligand Glycerol, (propane-1,2,3-triol) was obtained for further analysis. Similar structure search of different chemical compounds was done which were related to Glycerol, (propane-1,2,3-triol). At the end, 90 structures were obtained with their physical and chemical properties. Further, docking and screening were performed. ADMET study was done at the end of ADMET analysis and forty Chemical compounds were found to affect protein target.
Further, all parameters of those forty chemical compounds were analyzed and following twelve chemical compounds confirm their potential as drug target candidates. METHANOL – (DB00825),1,3BUTANEDIOL – (DB02202), CHOLICACID – (DB02659), LAURIC ACID – (DB03017), STERIC ACID – (DB03193), Aetiocholanolone– (DB02854), TRICOSANIC ACID – (DB03500), DEOXYCHOLIC ACID – (DB03619), Chenodeoxycholic acid– (DB06777), Stanolone – (DB02901), HYDEOXYCOLIC ACID – (DB11789), MESTEROLONE – (DB13587). From the above twelve compounds these three are hormone derivatives: Aetiocholanolone– (DB02854), Stanolone – (DB02901), MESTEROLONE – (DB13587).
Thus, it can be concluded that above twelve chemical compounds show great affinity against Dengue Virus Protein NS3 Helicase in complex with ssRNA. Hence these twelve chemical compounds can be further analyzed as drug target candidates for dengue fever disease in wet lab for the Production of new synthesis of drug against dengue fever disease.
ACKNOWLEDGEMENT
The authors are thankful to the Department of Biotechnology, Hemchandracharya North Gujarat University, Patan.
Conflict of Interest Statement
We, the authors of the submitted manuscript declare that the work and data present in the manuscript entitled – Design and prediction of Absorption, Distribution, Metabolism, Excretion and Toxicity properties of drugs for dengue fever using in-silico approaches is genuine research carried out by us. The work finally belongs to the institutes. We have not misused the data previously published and have not manipulated the original work.
References
Ackermann, M., & Padmanabhan, R. (2001). De Novo Synthesis of RNA by the Dengue Virus RNA-dependent RNA Polymerase Exhibits Temperature Dependence at the Initiation but Not Elongation Phase The Journal of Biological Chemistry, 276(43), 39926–39937. doi:10.1074/jbc.
Ago, H., Adachi, T., Yoshida, A., Yamamoto, M., Habuka, N., Yatsunami, K., & Miyano, M. (1999). Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure, 7(11), 1417–1426.
Alvarez, D. E., Laura, A., Ezcurra, D. L., Fucito, S., & Gamarnik, A. V. (2005). Role of RNA structures present at the 3 V UTR of dengue virus on translation , RNA synthesis , and viral replication. Virology, 339, 200–212. doi:10.1016/j.virol.2005.06.009
Bissantz, C., Folkers, G., & Rognan, D. (2000). Protein-Based Virtual Screening of Chemical Databases . 1 . Evaluation of Different Docking / Scoring Combinations. J. Med. Chem., 43, 4759–4767.
Bollati, M., Alvarez, K., Assenberg, R., Baronti, C., Canard, B., Cook, S.Bolognesi, M. (2010). Structure and functionality in flavivirus NS-proteins : Perspectives for drug design. Antiviral Research, 87(2), 125–148. doi:10.1016/j.antiviral.2009.11.009
Dengue Bulletin. (2016). World Health Organization, 39(December).
Dias, R., Filgueira, W., & Jr, D. A. (2008). Molecular Docking Algorithms. Current Drug Targets, 9, 1040–1047. doi:10.2174/138945008786949432
Fernando, L., Macedo, S., Pauli, I., Caceres, R. A., Filgueira, W., & Jr, D. A. (2008). Drug-Binding Databases. Current Drug Targets, 9, 1092–1099.
Iskar, M., Zeller, G., Zhao, X., Noort, V. Van, & Bork, P. (2011). Drug discovery in the age of systems biology : the rise of computational approaches for data integration. Current Opinion in Biotechnology, 23, 1–8. doi:10.1016/j.copbio.2011.11.010
Kuhn, R. J., Zhang, W., Rossmann, M. G., Pletnev, S. V, Corver, J., Lenches, E., … Lafayette, W. (2002). Structure of Dengue Virus : Implications for Flavivirus Organization , Maturation , and Fusion. Cell, 108, 717–725.
Mazzon, M., Jones, M., Davidson, A., Chain, B., & Jacobs, M. (2009). Dengue Virus NS5 Inhibits Interferon- a Signaling by Blocking Signal Transducer and Activator of Transcription 2 Phosphorylation. The Journal of Infectious Diseases, 200, 1261–1270. doi:10.1086/605847
Modis, Y., Ogata, S., Clements, D., & Harrison, S. C. (2003). A ligand-binding pocket in the dengue virus envelope glycoprotein. PNAS, 100(12), 6986–6991.
Mukhopadhyay, S., Kuhn, R. J., & Rossmann, M. G. (2005). A structural perspective of the. Nature Reviews, 3(January), 13–22. doi:10.1038/nrmicro1067
Qaddir, I., Rasool, N., Hussain, W., & Mahmood, S. (2017). Computer-aided analysis of phytochemicals as potential dengue virus inhibitors based on molecular docking , ADMET and DFT studies. J Vect Borne Dis, 4(September), 255–262.
Qi, R., Zhang, L., & Chi, C. (2008). Biological characteristics of dengue virus and potential targets for drug design. Acta Biochim Biophys Sin, 40 (2), 91–101. doi:10.1111/j.1745-7270.2008.00382.x
Song, C. M., Lim, S. J., & Chuantong, J. (2009). Recent advances in computer-aided drug design. Briefings in Bioinformatics, 10(5), 579–591. doi:10.1093/bib/bbp023
Wadood, A., Ahmed, N., Shah, L., Ahmad, A., Hassan, H., & Shams, S. (2013). In-silico drug design : An approach which revolutionarised the drug discovery process In-silico drug design : An approach which revolutionarised the drug discovery process. OA Drug Design & Delivery, 1(1), 1–4. doi:10.13172/2054-4057-1-1-1119
Whitehead, S. S., Blaney, J. E., Durbin, A. P., & Murphy, B. R. (2007). Prospects for a dengue virus vaccine. Nature Reviews, 5, 518–528. doi:10.1038/nrmicro1690
Williamson, E. M. (2014). Synergy : Interactions within Herbal Medicines Synergy and other interactions in phytomedicines. Phytomedicine, 8(5), 401–409.
Wilschut, I. A. R. J. (2010). Dengue Virus Life Cycle : Viral and Host Factors Modulating Infectivity Dengue virus life cycle : viral and host factors modulating infectivity. Cell. Mol. Life Sci., (67), 2773–2786. doi:10.1007/s00018-010-0357-z
Yang, J., & Chen, C. (2004). GEMDOCK : A Generic Evolutionary Method for Molecular Docking. Proteins: Structure, Function, and Bioinformatics, 55, 288–304. doi:10.1002/prot.20035


