1,2,3Department of Electronics and Communication Engineering, Sathyabama Institute of Science and Technology, Chennai. Tamilnadu. India.
Corresponding author email: srilatha169@gmail.com
Article Publishing History
Received: 15/04/2020
Accepted After Revision: 20/06/2020
The uncontrolled growth of cells in the brain causes brain tumour, there are two different types of tumours : malignant and benign. Malignant brings the cancerous type and spreads to other regions. At present, processing and developing of medical images is an important field. It categorises many types of imaging methods. Some of them are Magnetic Resonance Imaging (MRI), X- rays and computed Tomography scans
(CT scans) and so forth. The important goal of medical image processing is to identify accurate and meaningful information using images with the minimum error possible.MRI has high resolution and better quality images compared with other imaging techniques. So that, this technique is used to detect cancer tissues and to scan any part of the body. In this paper, neural network method is used for segmenting the brain tumours. The images were pre-processed for removal of noise and contrast-enhanced for better image. Accuracy, PSNR, Sensitivity were parameters calculated for the analysis of image quality.
Brain Tumour, Magnetic Resonance Imaging (MRI), X- Rays, Computed Tomography scans (CT scans), Malignant and Benign Tumour, Image Processing.
Srilatha K, Reddy C. S, Aravind C. Taxonomy of Medically Important Bacterial Species Through Intelligent Neural Network: A Soft-Computing Based Approach. Biosc.Biotech.Res.Comm. 2020;13(2).
Srilatha K, Reddy C. S, Aravind C. Taxonomy of Medically Important Bacterial Species Through Intelligent Neural Network: A Soft-Computing Based Approach. Biosc.Biotech.Res.Comm. 2020;13(2). Available from: https://bit.ly/3c1XLWR
Copyright © Srilatha et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
The uncontrolled growth of cancerous cells in the brain causes brain tumours. Tumours are classified as two types, they are malignant and benign. Malignant brings the cancerous type and spreads to other regions of the brain. The gliomas and meningioma are the examples for benign tumour and
astrocytma and glioblastomas are high level tumours classified as malignant type. According to the World Health Organization (Loius et al., 2018 WHO,2018), brain tumours are graded from grade I to grade IV to classify them as benign or malignant type. Usually, benign tumours fall under grade I and grade II and Grade III to grade IV glioma are classified as malignant type. All types of brain tumour produce symptoms like seizures, headache, problem with vision and hearing. Early diagnosis and treatment of tumour is very important, Due to the complex structure of the brain, detection and extraction of tumour is very important. There is a revolutionary development in the field of Medical Image analysis and computer vision within the past two decades. CT scans and MRI are used for the diagnosis of brain tumours,( Szil´agyi,.et al., 2018, Akter et al., 2018, Al-Ashwal et al., 2018).
This advance technology in the field of medical image processing can be used for segmentation of brain tumours. Brain tumour segmentation can be done by computer-aided systems for further medical analysis. Based on region, contrast, texture and colour the image is divided and it is called as segmentation. In this residual neural network method is used for segmentation of brain tumours, (Srilatha et al, 2019).
Brain MRI images with tumours like glioblastoma, cerebral metastasis, medulloblastoma were considered for segmentation. Mostly images are treated as two-dimensional signals and for manipulation processing techniques are applied. MATLAB, WINGRASS, and ArcGIS, MathCAD are the image-based analysis research software’s used to process images and to make an analysis on it, Kilic, et al, (2018).
A multi-paradigm numerical computing environment and proprietary programming language are the product of works in MATLAB. When images are as twodimensional data, this matrix laboratory is a useful tool for making analysis over images. Medical image processing is able to view the internal structure of the hidden organs in the body which are covered by skins and bones to predict the exact cause of the disease for further treatment procedure. The biological image is obtained from radiology (Ferlay et al, 2008), which uses technologies like X-ray, radiography, magnetic resonance imaging medical ultrasonography and endoscopy.
Srilatha. et al, (2019) and Al-Ashwal et al (2018) have proposed a system for early detection of brain tumour with image segmentation technique. The system uses a K-means clustering technique integrated with Fuzzy C means algorithm for brain MRI segmentation. They use two further segmentation such as threshold and level set segmentation to make an accurate brain tumour diagnosis. The system provides improved accuracy in various datasets.
Louis et al (20018) have proposed research on two-stage authenticated technique for detection of brain tumour. Right now, proposed system is a validated plan for the detection of a brain tumour, called a watershed matched methodology.The segmentation of brain tumour area was done
through classification approach of the watershed algorithm. In addition, scale invariant feature transform technique was also used to extract feature regions and matching divided area of brain tumour with an actual picture.
Kilic, et al, (2018) have carried research on the detection and visualization of the tissues of the brain. Detect the dimension of the tumour of the brain image, different stages of the brain can be determined. The step may be cancerous or noncancerous. To perceive the size of the brain, a tumour of
the brain can be recognized utilizing a k means clustering algorithm dependent on the detachment and morphological procedure. The morphological method used for the extraction of the features of the parts of the body through MRI scanning. Experimental analysis on calculating the extracted features of the portion of the brain disease.
Akter et al, (2018) have proposed a novel distributed picture segmentation technique for magnetic resonance brain pictures. In this technique, the different conversion is called a contour transformation that is the through canny edge detection. In this research work, the distribution technique
acquired for the improved method on contour constants through the canny edge detection approach. Experimental analysis was done on improvement of contour let conversion through the segmentation process using canny edge detection technique.
Melissa and Srilatha, (2016) presented a novel fuzzy c mean clustering approach that is also known as possible fuzzy c mean clustering technique. They separate of the database through middle free fuzzy c mean clustering into fuzzy c means clustering technique. The proposed method is less
prone to interference. In this research, it was analysed that the proposed method helps in the segmentation of the magnetic resonance imaging brain pictures along with intervention.
Szil´agyi et al., (2011, 2018) proposed segmentation of brain tumour from MRI using fast marching method. This method derives the gradients based on weights of each pixel in an image. The segmented image is done by watershed algorithm. This method acquires accuracy of 98.83%. Hamamci et al, (2012) and Akers et al, (1978) had proposed a method of segmentation of brain tumour using watershed technique and self-organizing maps. In this paper the brain images were skull stripped to eliminate the non-cerebral region, stationary wavelet transform (SWT) was used for feature extraction .then the tumour region was segmented using the watershed algorithm. This method combined with self-organizing maps achieved a
segmentation accuracy of 95.93%.
MATERIALS AND METHODS
The Block diagram shown in figure 1 represents the methodology for brain tumour segmentation. In the proposed work, The images were initially considered and tumour part is detected. Before segmenting the image, image is pre-processed by using gray scale, noise removal and CLAHE enhancement techniques. It is followed by the segmentation residual neural network technique and the performance analysis like accuracy, sensitivity and PSNR were calculated.
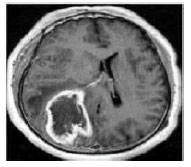
Data Search: Select an input image from data base or folder using Matlab. The selected input may be a colour image or binary image FIGURE.1.
Figure 1: Brain Tumour Input Image
Pre-Processing: The MRI images are altered by the bias field distortion. This makes the intensity of the same tissues vary across the image. Pre processing mainly focus on removing the noise present in the selected image. Conversion of the selected image is to gray scale representation is an important process in the technique. In gray scale the 3D image is converted to 2D image for fast process. The pictures were resized to 512×512 pixels.
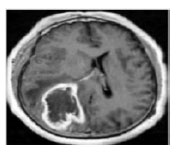
Removal of Noise In pre-processing noise is to be created in considered image. To remove that noise we are using Median filter. For digital image processing this Median filter is widely used because, It preserves the edges while removing noise, under certain conditions. The idea of using median filter is to run through the single entry by entry, replacing each entry with the median of neighbouring entries. patterns of the neighbours is known as “Window”, which slides entry by entry over the entire signal for 1D signals, the most obvious window is just the few preceding and following entries, where for 2D data the window must include all entries within a given radius shown in FIGURE.2.
Figure 2: Brain Tumour Pre-processing Image
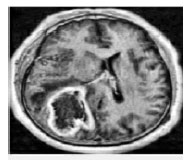
CLAHE (Contrast Limited Adaptive Histogram Equalization): In Contrast Limited Adaptive Histogram Equalization is utilized to enhance the contrast of the image. CLAHE works on little districts in the picture, called tiles, instead of the whole picture. Each tile’s difference is upgraded, with the goal that the histogram of the yield area around matches the histogram indicated by the ‘Appropriation’ parameter. The neighbouring
tiles are then joined utilizing bilinear shown in FIGURE.3.
Figure 3: Brain Tumour CLAHE Image
Residual Neural Network Segmentation: A Residual Neural Network is nothing but Artificial neural network of a kind that builds on constructs know from pyramidal cells in the cerebral cortex. This Neural Network work by utilizing skip connections, or shortcuts to jump over some layers. Extra weight matrix might be utilized to become familiar with the skip weights, these models are well known as Highway Nets.
In the context of residual neural networks, a non-residual neural network may be described as aplain network. In Residual Neural network the are three main layers they are Input Layer: The functionality of these layers are , where the images first entered into input layer after doing all function process like conversion of image in to grey scale, filtering, CLAHE for improve resolution of the image. Hidden Layer: After the image passes into next layer called hidden layer, the segmentation process done under this layer. The hidden layer consists many processing techniques. These processes can be done layer by layer Input layer: This image is put in this layer.
Binary Convolution layer: it comprises in including a sign or a picture with piece to get highlight maps. Thus, a section in a component map is connected to the past layer through the loads of the parts. The loads of the parts are adjust during the preparation stage by back proliferation, so as
to upgrade certain attributes of the info. Since the bits are shared among all units of the same feature maps, Convolutional layers have fewer loads to prepare than thick FC layers, making simpler to prepare and less inclined to over fitting. In addition, since a similar portion is convolved over the whole picture, a similar element is recognized freely of the area interpretation in variance.
Activation Function: it is liable for Non- Linearly transforming the information.
Rectifier linear units (ReLU) defined as,
f(x) = max(0, x)
Were found to accomplish preferred outcomes over the more traditional sigmoid, or hyperbolic digression capacities, and accelerate preparing. Be that as it may, forcing a steady 0 can impede the angle streaming and subsequent modification of the loads. We adapt to these confinements utilizing a variation called flawed rectifier direct unit (LReLU) that presents a little incline on the negative piece of the capacity. This capacity is characterized as Where α is the brokenness parameter. In the last FC layer, we utilize delicate max. Pooling: It consolidates spatially close by features in the component maps. This combination of possibly redundant features makes the representation more compact and invariant to small image changes, such as insignificant details; it also decreases the computational load of the next stages. The common pooling techniques to use maxpooling or average-pooling. In max-pooling the maximum pixel is taken into account and remaining all is removed.
Training: To train the RNN the loss function must be minimized, but it is highly nonlinear. We utilize Stochastic Slope Drop as a streamlining calculation, which makes strides relatively to the negative of the angle toward neighbourhood minima. All things considered, in districts of low ebb and flow it tends to be moderate.
In this way, we utilized Nesterov’s Quickened Force to quicken the calculation in those areas. The force ν is kept consistent, while the learning rate ε was straight diminished, after every age. We consider an age as a total ignore all the preparation tests. Residual layer: In this based on their weights the image pixels is skipped. If layers are not necessary we can skip and reduce the time of execution. This is very important layer in this segmentation process shown in FIGURE.4.
Figure 4: Block diagram of Proposed Methodology
The process technique layers in the hidden layer are repeated until the image is extracted perfectly. Output Layer: The segmented image is
shown is the output layer. The tumour detected image is compared with all the existing methods.
RESULTS AND DISCUSSION
In this paper Neural Network Method is used for segmentation of brain tumour by considering the gradient weight of each pixel. The MRI images of brain are taken from radiopedia. The MRI images of the brain with Glioblastoma, cerebral metastasis and meningioma are considered for research
purpose. The performance analysis is calculated based on the segmented image. The calculated values are to be compared with existing methods.
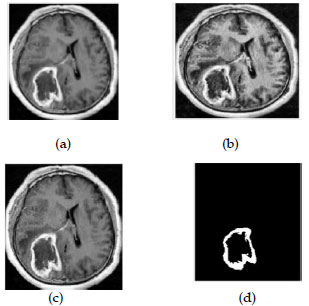
FIUGURE.5. (a)input image(b)filtered image(c) enhanced image with seed point(d)Tumour segmented and extracted image.
Figure 5: Brain Tumour Identification and Detection Image (a-d)
This process mainly focused to correct the intensity of t he image and removing the noise of the image. The above figures shows that how the segmented image is extracted from the input image. The filtered image removes the noise from the original image and enhanced image for the resolution of image .The RGB colour image for displaying the segmentation part of the tumour image. This RGB colour model used for MRI scanned images to display the result of the tumour. Bar Chart and Comparison Graph shown in Fig.6.
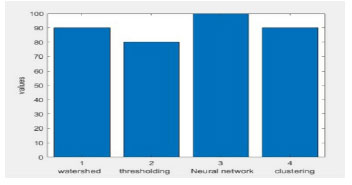
Figure 6: Brain Tumour comparison with various methods Accuracy bar chart
The above graph shows that accuracy values in the existing methods and the proposed methods and specificity has been calculated and compared with other methods. The highest and lowest accuracy values of the tumour images are displayed. The comparision of accuracy, sensitivity and
Specificity values. The accuracy and specificity can be calculated by following equations.
Accuracy=(TP+TN)/(TP+TN+FP+FN)
Specificity=TN/(TN+FN)
Sensitivity=TP/(TP+FN)
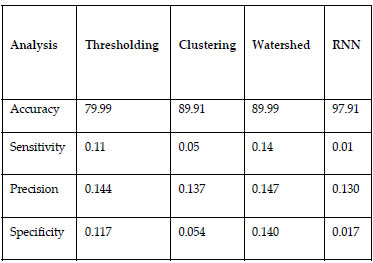
The below TABLE.1. shows that comparison of existing and proposed methods for different parameters. It also displays high accuracy values and
specificity values to identify tumour.
Table 1. Performance evaluation of segmentation
CONCLUSION
The proposed system is used to detect brain tumour using residual neural network segmentation technique. The proposed method produces exact location of the tumour in the image. To get an MRI input image a GUI is created , pre-processed which includes conversion of gray scale , filtering of image using median filter to remove noise ,enhancement can be done using CLAHE for resolution of image and segmentation using residual neural network technique. Then the results were analysed. The median filter and CLAHE also employed to get clear image of brain affected by tumour. The residual neural network segmentation used for layerization of every pixel of images. The accuracy is analysed in terms of PSNR, sensitivity and specificity. This segmentation reduces the error rate of MRI brain image. The number of layers used in this neural network is less and executes fast. The accuracy values also high compared to other techniques.
REFERENCES
Akers SB, (1978) Binary decision diagrams IEEE Transactions on Computers, vol. C-27, pp. 509–516
Akter, L. A., and Kwon, G. R. (2018). Integration of Contourlet Transform and Canny Edge Detector for Brain Image Segmentation. In 2018 Tenth International Conference on Ubiquitous and Future Networks (ICUFN) (pp.798-800). IEEE.
Al-Ashwal RH, Eko Supriyanto, Nur Anati BAbdul Rani, Nur Azmira, B Abdullah, Nur IllaniB Aziz, Rania B Mahfooz,(2018) Digital Processing of Computed Tomography Images:Brain Tumour Extraction and Histogram Analysis, Mathematics and Computers in Contemporary Science, ISBN 978-960-474- 3568.
Bauer, S., Wiest, R., Nolte, L.P. and Reyes, M.(2013), A survey of MRI-based medical image analysis for brain Tumour studies. Physics in Medicine & Biology, 58(13), p.R97.
Eman Abdel-Maksoud, Mohammed Elmogy, Rashid Al-Awadi (2015) Brain tumour segmentation based on a hybrid Clustering technique Egyptian Informatics Journal, Elsevier 2015.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C and Parkin DM,(2011) GLOBOCAN v2.0, Cancer Incidence and Mortality Worldwide International Agency for Research on Cancer, Lyon, France, 2010.http://globocan.iarc.fr, Accessed on November 13, 2011 .
Hamamci A., N. Kucuk, K. Karamam, K. Engin and G. Unal, (2012)TumourCut: segmentation of brain tumours on contranst enhanced MR
images for radiosurgery applicarions, IEEE Trans. Med. Imag., vol. 31, pp. 790–804, 2012.
Kilic, H., Yuzgec, U., & Karakuzu, C. (2018). A novel improved antlion optimizer algorithm and its comparative performance. Neural Computing
andApplications, https://doi.org/10.1007/ s00521-018-3871-9,2018.
Liu Y, Y. Liu, H. Xu, and K. L. Teo, ( 2018) Forest fire monitoring, detection and decision making systems by wireless sensor network, in 2018 Chinese Control And Decision Conference (CCDC), 2018, pp. 5482- 5486.
Louis D.N., Ohgaki H., Wiestler O.D, Cavenee (2018) (Eds.), WHO Classification of Tumours of the Central nervous. Melissa, S., Srilatha, K.(2016) A novel approach for pigmented epidermis layer segmentation and classification, International Journal of Pharmacy and Technology, Volume 8(1), pp-10449-10458.
Njeh I, L. Sallemi, I. Ben Ayed, K. Chtourou, S. Lehericy, D. Galanaud and A. Ben Hamida, (2015) 3D multimodal MRI brain glioma tumour
and edema segmentation: a graph cut distribution matching approach, Comput. Med. Imag. Graph., vol. 40, pp. 108–119
Naresh Karthik MD, I Kathirvelan, K Srilatha, (2017) Survey on Image Segmentation in Various Clustering Algorithms, March – April RJPBCS 8(2) pp. 1131-1136
Paolo E. Santangelo , Bryson C. Jacobs , Ning Ren : (2014)Suppression effectiveness of watermist sprays on accelerated wood- crib fires, Fire Safety Journal 70 (2014) 98111
Raza U, P. Kulkarni, and M. Sooriyabandara.: (2017) Low power wide area networks: An overview. IEEE Communications Surveys
Tutorials, 1–1
Redmon J, Farhadi A (2018) N YOLOv3: An Incremental Improvement[J].pp.1-6, https://arxiv.org/abs/1804.02767, 2018.
Srilatha, K.,Ulagamuthalvi, V, A (2019) Comparative Study On Tumour Classification, Research Journal Of Pharmacy And Technology, Volume 12 (1),pp-407-411
Szil´agyi L, S. M. Szil´agyi, and B. Beny´o, (2012) Efficient inhomogeneity compensation using fuzzy c-means clustering models”,
Comput. Meth. Progr. Biomed, vol. 108, no. 1, pp. 80–89, 2012.
Szil´agyi, L D. Icl˘anzan, Z. Kap´as, Zs. Szab´ o, ´A. Gy˝orfi, and L. Lefkovits,(2018) Low and high grade glioma segmentation in multispectral
brain MRI data, Acta Universitatis Sapientiae, Informatica, vol. 10, no. 1, pp. 110–132
Szil´agyi, LD. Lefkovits L and B. Beny´o,(2015) Automatic brain tumour segmentation in multispectral MRI volumes using a fuzzy cmeans
cascade algorithm Proc. 12th International Conference on Fuzzy Systems and Knowledge Discovery (FSKD 2015, Zhangjiajie, China), pp. 285-291
Szil´agyi, S. M. Szil´agyi, B. Beny´o and Z. Beny´o, (2011) Intensity inhomogeneity compensation and segmentation of MR brain
images using hybrid c-means clustering models Biomed. Sign. Proc. Contr., vol. 6, no. 1, pp. 3– 12
Tustison NJ , B. B. Avants, P. A. Cook, Y. J. Zheng, A. Egan, P. A. Yushkevich and J. C. Gee (2010) N4ITK: improved N3 bias correction,”
IEEE Trans. Med. Imag., vol. 29, no. 6, pp. 1310–1320, 2010.
Vovk U, F. Pernu˘s and B. Likar,(2007) A review of methods for correction of intensity inhomogeneity in MRI IEEE Trans. Med. Imag., vol. 26, pp. 405–421