1Department of Microbiology, Sri Sarada Niketan College of Science for Women, Karur, Tamil Nadu, India.
2Post Graduate and Research Department of Microbiology, Hindustan College of Arts and Science, Tamil Nadu, India.
Corresponding author email: chithumicros@gmail.com
Article Publishing History
Received: 25/09/2021
Accepted After Revision: 22/12/2021
Oil bio-degradation mechanism by microorganisms is requested for an effective microbial remediation of soil contamination by oil spills. The current examination pointed the identification of a biosurfactant producing bacteria for biosurfactant production from oil contaminated sites from Tamil Nadu. The biosurfactant testing screening methods were used to screen the potent strains and sequencing studies were used for Pseudomonas species identification. The bacterial isolate BS17 subjected to be the potent enzyme (Protease, Lipase and Esterase) producer. Among the tested production media, the ground nut oil cake was identified to be the optimum media for protease (0.47069 Unit/ml), lipase (9 Unit/ml) and esterase activity (3.891 Unit/ml) for bacterial isolate BS17.
The bacterial isolate BS17 showed greatest lipase (15 Unit/mL) protease (0.8067 Unit/mL) and esterase (4.756 Unit/mL) enzyme activity at pH 9.0. At 35 ℃ bacterial isolate BS17 showed greatest enzyme action in protease (1.2772 Unit/mL), lipase (17 Unit/mL) and esterase (5.2972 Unit/mL) enzyme activity. At 48hrs of incubation period bacterial isolate BS17 showed most extreme enzyme activity in protease (3.361 Unit/mL), lipase (28 Unit/mL) and esterase (8.918 Unit/mL). The sequence of BS17 was deposited in NCBI and Accession number was received [MT337593.1]. Statistical analysis with the minimum significant difference (LSD) test of ANOVA was carried out to determine the oil degradation efficiency. This paper demonstrated the isolated P. aeruginosa (BS17) crude oil biodegradation from oil contaminated land soil sample. Strain BS17 was proved as potent bio-surfactant producer using crude oil by utilizing carbon and energy source in oil degradation mechanism.
Esterase, hydrocarbons, lipase, protease, P. aeruginosa, sequencing.
Chithra S, Hemashenpagam N. Biosurfactant-Producing Bacteria Isolated from Oil Ontaminated Soil and Its Media Optimization for Enzyme Production. Biosc.Biotech.Res.Comm. 2021;14(4).
Chithra S, Hemashenpagam N. Biosurfactant-Producing Bacteria Isolated from Oil Ontaminated Soil and Its Media Optimization for Enzyme Production. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/3qEpNSf“>https://bit.ly/3qEpNSf</a>
Copyright © Chithra and Hemashenpagam This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Soils contaminated by petrol hydrocarbons can influence soil well-being. Also, increased focuses on the consequences for human health well-being. They can kill soil microorganisms, decreasing their number and bio-activity. Soil organisms help make supplements accessible to plants. ‘Reducing microorganisms’ numbers or action additionally influences plants. Some petrol hydrocarbons can be taken up by plants and represent a danger to grazing animals, wildlife, and plant-eating bugs.
With a significant level of petroleum contamination, seeds can’t grow. At times, even develop plants can’t grow any longer. Some oil-based goods can extremely clog the soil and so it affects the penetration of water into the soil. Polycyclic aromatic hydrocarbons (PAHs) are harmful impurities in different conditions since it has cancer-causing and mutagenic impacts (Silva et al. 2020; Patel et al. 2020).
The biodegradation of PAHs can be considered on one hand to be a piece of the typical cycles of the carbon cycle, and then again as the expulsion of man-made poisons from the climate. The utilization of microorganisms for biodegradation of PAH-contaminated soil and other source of petroleum contaminated environments.
Among toxins of hydrocarbons, other petroleum source (alkanes and aromatic compounds combination) is reported as soil hydrocarbon substance leakage from ship tanks or re-rented in unintentional leakage accident. Hydrocarbons are for the most part unmanageable and impervious to microbial attack because of their low dissolvability and bioavailability. In this way, these mixtures are profoundly industrious in the climate and are bio-accumulated in microorganisms (Azadi et al. 2020).
The accomplishment of bio-degradation is reliant upon the microbial capacity to degrade the hydrocarbons and their growth and metabolic growth kinetics. Higher rates of hydrocarbon degradation are regularly accomplished with a bacterial enhancement consortium confined from the climate that necessities bio-restoration. To get efficient diesel oil-degrading bacterial cultures, information on the variety of the microbial local area presents in polluted soil with diesel oil, their metabolic highlights, and their ability to degrade crude oil.
The important properties of hydrocarbon-degrading bacterial species are the production of bio-surfactants (Habib et al. 2020). These organisms produce small surfactant atoms that decrease surface tension. Bacterial consortia often reported as efficient degrader of hydrocarbons and emulsifying compound production. The application of bio-surfactants activating and eliminating the impurities by solubilization and emulsification in waste treatment (Mujumdar et al. 2019; Lawniczak et al. 2020).
There are various studies reported on bacterial bio-surfactants, but the potency of activity is ——–based on their compound chemical nature. A Pseudomonas species was known for biosurfactant production. Microorganisms produce biosurfactants as biofilm which associates with an interface and modifies the surface properties like wettability and different properties. A bacterium isolated from sea water polluted with oil, Pseudomonas aeruginosa has proved the capacity to breakdown hydrocarbons following 28 days of incubation.
The percentage of degradation by the above species has been studied because of bio-surfactant production. Bioremediation is the main technique that has been acknowledged treatment by utilizing native microbial colonies. Certain bio-surfactant-producing microorganisms can degrade a few classes of hydrocarbons. Bio-surfactant characterization is classified based on the microbes’ type, compounds nature, the polar groups of compounds. Bio-surfactants are reported as one of the non-toxic substances, as they are one of the good choices in cosmetic, food, and pharmaceutical industries (Lawniczak et al. 2020).
In this work, we have collected oil spilled soil samples from various locations and bacteria cultures were isolated to test the efficiency of bio-surfactant production. In recent research papers, the bacteria belong to 79 genera are reported as petroleum hydrocarbon degraders which includes Pseudomonas have been observed for bio-degradation of crude oil (Ma et al. 2021). Our study reported that isolated strain is a potent strain to facilitate enzyme production in a large-scale up process. Moreover, this strain is proved with higher efficiency in bio-surfactant production.
MATERIAL AND METHODS
General microbiological and laboratory techniques for the readiness of media, bacteria isolation, and stock culture were maintained. Land oil spilled soil was collected from distinct districts of Tamil Nadu. The examples were collected at a profundity of 5cm from the outside of the dirt. By following serial dilution and streak plate method, the bacterial colonies were isolated and slanted cultures was prepared to maintained the stock culture for further examination. The isolated bacterial colonies were inoculated in nutrient broth with the addition of 0. 1 gram of oil (engine oil, petrol, or diesel) and incubated the culture for 2 days.
After incubation, the culture was centrifuged for 12 min at 4500 rpm speed to remove the bacterial cells. After centrifugation, the cleared layer of supernatant was carefully transferred into a new tube and used the same for further characterization studied. A 10 μl of the crude bio-surfactant was spotted on a thin layer chromatography plate in mobile phase of chloroform, water, and chloroform (CHCl3: H2O CH3OH) in the ratio of 70:0. 5:10 (v/v/v) as a mobile phase. The developing reagent (ninhydrin) was utilized to visualize lipid-based bio-surfactant (red spot) and anthrone reagent to visualize glycol-based lipid bio-surfactant (yellow spot). Rf values were then calculated (Rani et al. 2020).
Drop collapse test was examined to screen the bio-surfactant-producing strain. It relies upon the rule that a dropped of fluid containing a bio-surfactant would fall totally over the oil surface. 4μl of mineral oil was added to a cleared glass slide and incubated the slide at room condition and afterward, 6 μl of supernatant was applied to the glass slide with oil covered, and size of each dropped was seen. The outcome was viewed as sure when the width of the dropped was expanded by 1mm from that which was formed by deionized watered which was taken as the negative controlled (Rani et al. 2020).
For the emulsification stability test, E24 of bacterial cultures was dictated by the addition of 3 ml oil to a similar measured of culture tube, following 3 min shaking in vortexer and kept undisturbed for 1 day (Rani et al. 2020). The percentage of emulsification capacity was determined:
Emulsification stability % = (Total Height of the emulsified layer / height of total liquid column) ×100
For the lipolytic activity, about 1 gm of agar and gelatin was weighed and dissolved in 100 ml of deionized watered and autoclaved. About 25 ml of the above mixture was poured into a sterile petri plate and wells was cut and the supernatant containing bio-surfactant was added into the wells with a positive controlled under aseptic conditions and incubated at room temperature for one day. Following that the plates was developed with 15% mercuric chloride prepared in 6M HCL solution and the mean diameter was noted. Lipase-producing bacterial strains formed a zone of cleared hollow (hydrolysis) on the tributyrin agar medium.
The color zone was estimated after 24 hours of incubation at 37°c. The lipolytic activity of each bacterial culture was determined by estimating the diameter of the zones. The strain with the highest zone was utilized for additional examination. The media utilized contained peptone 1 gm, NaCl 0. 5 gm, CaCl2 2H2O 0. 01 gm, agar 1. 8 gm, pH 7. 4 for 100 ml media quantity, and sterilized. Tween 80 was included at a final working concentration of 1% (v/v). Then the isolates was spread on the plate, incubated for 24 hours, and the formation of color halos was noted. The strain with the biggest color halo zone was utilized for additional investigation (Guan et al. 2020).
For protease activity, 500 μl of casein (1 %), and 450 μl of 0. 2 mol/l phosphate buffer (pH 7. 0) was added to 350 μl of crude enzyme extract. At 60 °c the reaction content was incubated for 10 min. The reaction was stopped by the 2 ml of 10 % trichloroacetic acid addition. Then the tube was centrifuged at 7000 x g for 12 minutes and to the separated supernatant, 6 ml of 0. 3 mol/l Na2CO3, 2 ml of folin and ciocalteau’s phenol reagent was added. Followed by 30 min incubation and measured at 660 nm utilizing tyrosine as a standard, the absorbance of the blue color formed (Habib et al. 2020).
The free lipase activity was examined by spectrophotometrically by following the method with some modifications ((Kumar et al. 2020). The reaction mixture contains 0.05M phosphate buffer (pH 8. 0) 3 ml, 0.8 mM p-NPP (0. 1 ml), 0. 2 ml of lipase and incubated at room temperature. And by adding 1 ml of ethanol, the reaction was stopped. A controlled was studied without the addition of enzyme. Absorbance was measured at 410 nm using UV spectrophotometer. One international unit (IU) of lipase action was measured when the enzyme catalyzing and the released of 1 μmol of p-nitrophenol per min from p-NPP (Abi Habib et al. 2020).
For esterase activity, the assay solution contained 15 ml of 1% (v/v) tributyrin (substrate), 1% solution of gum acacia, 600μl calcium chloride (2%) and 1. 5 ml of NaCl (1M). The whole reaction mixture was studied by titration 10mM NaOH (10mM) and the esterase activity was calculated.
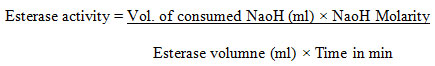
One unit of esterase activity was concluded as the amount of enzyme that released 1 μmol fatty acid min-1 at 30 ℃ with pH 7 under the test conditions (Sani et al. 2021). For the media optimization and to induce the maximum enzymatic activity of protease, lipase, and esterase, the parameters of the following factors like media, pH, temperature, incubation period, carbon source, and nitrogen source was optimized based on a one-factor at a time method. The bacterial isolates were cultured on different substrates (4% w/v) such as coconut oil cake, cottonseed oil cake, groundnut oil cake, gingelly oil cake, soya bean oil cake, rice bran, wheat bran, and the studied was compared with nutrient broth medium at 37 ℃ with pH of 7. 5 and incubated for 48 hours.
The cultured broth was then centrifuged at 12,000 rpm for 15 minutes. The cleared supernatant was collected and used as a crude enzyme for studying the enzymatic activity and total protein content. The effect of pH on enzymatic activity was examined with various pH 4 – 12. The ideal pH for maximum enzyme activity was noted by changing the pH of the medium. The effect of temperature on enzymatic activity was examined with different temperatures such as 25 ℃, 30 ℃, 35ºc, 40 ℃, 45 ℃, and 50 ℃.
The ideal temperature for maximum enzyme activity was chosen by changing the temperature of the medium. The effect of the incubation period on enzymatic activity was studied at different time intervals that include 12 hours, 24 hours, 36 hours, 48 hours, 60 hours, and 72 hours. The ideal incubation period for the highest enzymatic activity was recorded by changing the incubation period of the medium (Al-Dhabi et al. 2020).
For morphology study and molecular identification, the most effective bacterial culture was distinguished by standard biochemical tests and morphological examination by microscope. Genetic identification of strain in which the genomic DNA of the bacteria was extracted utilizing the standard procedure. Then the 16s rDNA was amplified used PCR by utilizing universal primer, 968F (AACGCGAAGAACCTTAC) and 1541R (AAGGAGGTGATCCAGCCGCA) (Fadeev et al. 2021). Polymerase chain reaction (PCR) was performed in a 30 μl volume of master mixed (sigma aldrich, USA) and 10 ng of genomic DNA.
PCR amplicons was further separated on 1 % of agarose gel and PCR amplicons (16s rRNA gene) was purified used pcr clean-up kit (genelute™). PCR amplicons was sequenced by a single by-pass sequencing method. The consensus sequence of 16s rDNA was utilized for basic local alignment searched tool (BLAST) examination against the data set in the National Center for Biotechnology Information (www. ncbi. nlm. nih. gov). Sequence alignment was done by utilizing Clustal w and by dictated by the neighbor-joining technique the phylogenetic tree was constructed used Mega 5 software (Modi et al. 2019). The curated FASTA sequence was submitted to Genbank and the accession number was acquired.
RESULTS AND DISCUSSION
All the dirt soil samples were gathered at a profundity of 5cm from the land surface. The gathered soil samples had an identification of dark tone because of consistent oil spillage. The gathered oil spilled soil samples in sterile polythene sacks were stored at 4°C aseptically and utilized for additional exploration studied. Most of the strains was belongs to Pseudomonas genus, especially P. aeruginosa species, which widely reported for crude oil degradation (Shweta et al. 2021).
Figure 1: Thin Layer Chromatography: TLC analysis of purified bs produced two spots in the TLC plate with an Rf value of 0.89. The bio-surfactant fraction after centrifugation showed a positive result with Molish reagent and vapor of iodine, representing the lipid moieties presence and carbohydrate moieties. The TLC analysis determined the presence of the glycolipid nature of extracted bio-surfactant.
Figure 1:

Oil spreading and Drop collapse test: Among the other selected bacterial isolates, the broth culture of bacterial isolate BS17 was noted as the highest oil spreading efficiency (38 mm) followed by bacterial isolate 45 (36 mm). The bacterial isolates 40 (7 mm), 41 (9 mm), 70 (4 mm), 62 (8 mm) were observed as the lowest zone formation. The remaining bacterial isolates observed have no zone of displacement which is recorded as an absence of bio-surfactant production. The bacterial isolate BS17 was observed as the highest bio-surfactant activity in the drop collapse test (Rani et al. 2020).
Emulsification stability test: The effectiveness of biosurfactants in supplying oil to bacterial cells was investigated by measured the emulsifying activity (E24 index) of the biosurfactant. The screening was studied by adding 2 ml of gasoline to 1 ml of the supernatant and stirring vigorously and keeping overnight. The emulsification activity was calculated as a percentage. Among the fifty bacterial isolates screened, the emulsifying activity was found have been maximal in bacterial isolate BS17 (55 %), followed by bacterial isolate BS56 (48 %). The lowest leveled of emulsion activity (0. 3 %) was observed in bacterial BS38, BS32, and BS45 (Rani et al. 2020).
Extraction of protease, lipase & esterase enzymes from supernatant containing bio–surfactant: From the past outcomes concerning the fundamental screening of potent bacterial isolates for the production of biosurfactants by oil spread test, drop collapse method, and emulsification activity test. Based on the result we got, 31 bacterial isolates was separated to assess the lipolytic, proteolytic, and esterolytic activities by estimating the hydrolysis of tributyrin, gelatin, and tween 80 around the bacterial colonies.
Out of the 31 bacterial isolates BS27 bacterial isolates was observed for lipolytic activity and isolate BS17 was identified have been the best lipase producer. While 24 bacterial isolates showed esterolytic activity and 25 bacterial isolates showed proteolytic action with bacterial isolate BS17 was showed have been the best esterolytic and proteolytic production. The bacterial isolates BS17, BS34, and BS47 showed the best lipolytic activity with no response of protease and esterase activity. Despite the reports that these enzymes were highly demanded for various applications in biotechnology industries (Al-Ghanayem et al. 2020).
Oil Degradation by extracted bio–surfactant: The measured of oil in contaminated soil samples before microbial treatment and after treatment with the particular bio-surfactants was assessed utilizing the gravimetric technique. In the supernatant of bacterial isolate BS56, the leveled of oil degradation percentage was 88. 75 %. This was supposed have been the highest degradation percentage among the screened 17 bacterial isolates. Followed that, the bacterial isolate BS17 showed an oil degradation percentage of 87. 4 1%. In the supernatant of bacterial isolate BS39, the degradation of oil percentage was 79. 30 %. This was recorded as the least oil degradation percentage (Christova et al. 2019).
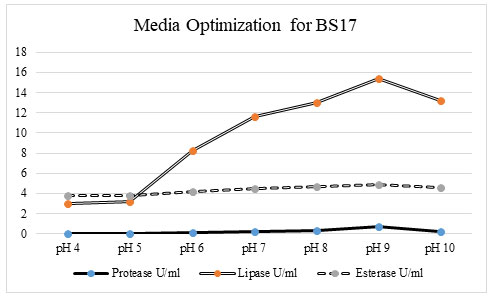
Optimization of media for promoting the enzyme activity: Production media plays a crucial role in the production of microbial enzymes. Different media such as coconut oil, cottonseed oil cake, groundnut oil cake, gingelly oil, soyabean oil cake, rice bran, nutrient broth, wheat bran was tested to studied the effect of media on maximum enzyme production for protease, lipase, and esterase activity. From the previous data we got, the bacterial isolate BS17 and isolates was selected for their ability to produced lipase, protease, and esterase in higher production. The bacterial isolates BS17 and was subjected have been the potent producer and was chosen for further optimization studied (Al-Dhabi et al. 2020).
Figure 2: Among the tested production media, the groundnut oil cake was found have been the optimum media for protease (0. 47069 u/ml), lipase (9 u/ml), and esterase activity (3. 891 u/ml) for bacterial isolate BS17.
Optimization of pH for promoting the enzyme activity: The role of pH was one of the significant variable parameters for microbial growth in the production medium. As the pH assumes a significant part in all the organic cycles, so the enzyme production was tested at various pH liked 4 – 12.
Figure 3: The investigation of the impact of various pH uncovers that enzyme production was extraordinarily affected by fluctuating pH of the production media. The bacterial isolate BS17 showed greatest lipase (15 U/ml) protease (0. 8067 U/ml) and esterase (4. 756 U/ml) enzyme activity at pH 9. 0. At pH 4. 0, 5. 0 and 12. 0 the bacterial isolate 17 recorded total shortfall of protease activity and low lipase activity (2 U/ml, 3 U/ml and 3 u/ml) individually and esterase action (3. 676 U/ml, 3. 837/U/ml and 4. 162 u/ml). As there was an increment in the pH esteem from pH 4. 0 to 9. 0 there was a significant increase in lipase, protease, and esterase production in bacterial isolate BS17. The protein content was also recorded high up to pH 9. 0 after which there was a rapid decrease in the protein content of bacterial isolate BS17 which relates to the decrease in enzyme production (Hou et al. 2021).
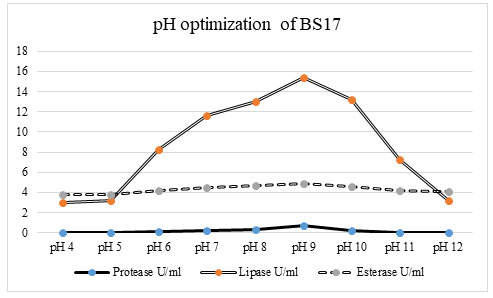
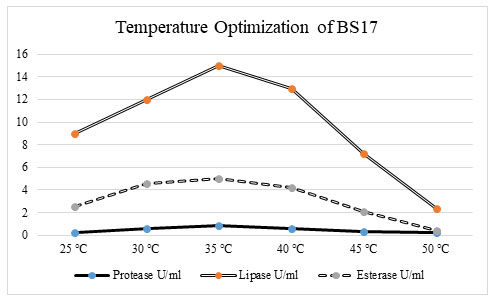
Figure 4: Optimization of temperature for promoting the enzyme activity. At 35 ℃ bacterial isolate BS17 showed the greatest enzyme action in protease (1. 2772 U/ml), lipase (17 U/ml), and esterase (5. 2972 U/ml) enzyme activity. There was an increase in chemical creation in protease, lipase, and esterase action when the temperature increments from 25 ℃ to 35 ℃. Continuously there was a decline in a compound movement when the temperature ascends from 40 ℃ to 50 ℃. The protein content additionally showed a significant increase up to temperature 35 ℃ after which there was a decrease in the protein content of bacterial isolate BS17 (Benabda et al. 2019).
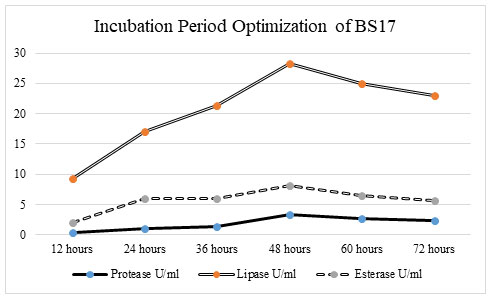
Optimization of incubation period on enzyme activity: Incubation time assumes a significant part in enzyme production. To examine the impact of the incubation period on protease, lipase, and esterase production, different incubation periods, such as 12 hrs, 24 hrs, 36 hrs, 48 hrs, 60 hrs, and 72 hrs was utilized.
Figure 5: At 48hrs of the incubation period, bacterial isolate BS17 showed the most extreme enzyme activity in protease (3. 361 U/ml), lipase (28 U/ml), and esterase (8. 918 U/ml). There was an increment in enzyme catalysis in protease, lipase, and esterase activity when the incubation time frame increases from 12 hrs to 48 hrs. There was a critical lessening in enzyme production of protease, lipase, and esterase when the incubation time frame ascends from 60 hrs to 72 hrs. Least protein content in protease (0. 4033 U/ml), lipase (9 u/ml) and esterase (2. 972 U/ml) action was recorded as 12 hrs incubation period. The protein content likewise showed a huge increase up to 48 hrs incubation period after which there was a significant reduction in the protein content of isolate BS17 (Chimbekujwo et al. 2020).
Molecular identification of potent bio–surfactant producing bacterial isolates by Phylogenetic tree construction:
Figure 6: The potent isolate BS17 has shown maximum response in all screening tests. The sequenced 16s rRNA was curated and the phylogenetic tree was constructed used Mega5 software to align the closely related species. From the analysis, the BS17 was closely related to Pseudomonas aeruginosa. The 16s rRNA sequence of BS17 was submitted and the NCBI Genbank accession number was received [MT337593.1].

Statistical analysis: Results and all the experiments were represented as standard deviation (SD) and ± mean. ANOVA table was used to determine the significant differences statistically (p < 0. 05) by used SPSS 16. 0 (IBM Corp, USA) (Mishra et al. 2019).
CONCLUSION
The findings of the present study suggests that the biodegradation of crude petroleum by P. aeruginosa BS17 isolates from hydrocarbon-contaminated soil. Strain BS17 was identifies to be an effective raw petroleum degrader and could potent biosurfactant utilizing raw petroleum as the sole carbon and fuel source throughout degradation. The biosurfactant produces by BS17 has high surface action and displays superb emulsification action against various hydrocarbon substrates. Every one of these positive properties works with the strain as a proficient bacterial biosurfactant in different ecological applications, especially in the remediation of unrefines petroleum pollutes areas.
Conflict of Interests: Authors have no conflict of interests to disclose.
REFERENCES
Abi Habib, J., De Plaen, E., Stroobant, V. et al. (2020). Efficiency of the four proteasome subtypes to degrade ubiquitinated or oxidized proteins. Sci Rep 10, 15765. https://doi.org/10.1038/s41598-020-71550-5.
Al-Dhabi, N. A., Esmail, G. A., Ghilan, A. M., et al. (2020). Isolation and screening of Streptomyces sp. Al-Dhabi-49 from the environment of Saudi Arabia with concomitant production of lipase and protease in submerged fermentation. Saudi journal of biological sciences, 27(1), 474–479. https://doi.org/10.1016/j.sjbs.2019.11.011
Al-Ghanayem, A. A., and Joseph, B. (2020). Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 104, 2871–2882. doi: 10.1007/s00253-020-10429-x
Ambust, S., Das, A.J. and Kumar, R., (2021). Bioremediation of petroleum contaminated soil through biosurfactant and Pseudomonas sp. SA3 amended design treatments. Current Research in Microbial Sciences, 2, p.100031., https://doi.org/10.1016/j.crmicr.2021.100031.
Azadi, D., and Shojaei, H. (2020). Biodegradation of polycyclic aromatic hydrocarbons, phenol and sodium sulfate by Nocardia species isolated and characterized from Iranian ecosystems. Sci Rep 10, 21860. https://doi.org/10.1038/s41598-020-78821-1
Benabda, O., M’hir, S., Kasmi, M., et al. (2019). Optimization of protease and amylase production by Rhizopus oryzae cultivated on bread waste using solid-state fermentation. Journal of Chemistry, 2019. https://doi.org/10.1155/2019/3738181
Chimbekujwo, K.I., Ja’afaru, M.I. and Adeyemo, O.M., (2020). Purification, characterization and optimization conditions of protease produced by Aspergillus brasiliensis strain BCW2. Scientific African, 8, p.e00398.
Christova, N., Kabaivanova, L., Nacheva, L., et al. (2019). Biodegradation of crude oil hydrocarbons by a newly isolated biosurfactant producing strain. Biotechnology & Biotechnological Equipment, 33(1), pp.863-872, DOI: 10.1080/13102818.2019.1625725
Fadeev, E., Cardozo-Mino, M.G., Rapp, J.Z., et al. (2021). Comparison of Two 16S rRNA Primers (V3–V4 and V4–V5) for Studies of Arctic Microbial Communities. Front. Microbiol. 12:637526. doi: 10.3389/fmicb.2021.637526
Guan, C., Tao, Z., Wang, L. et al. (2020). Isolation of novel Lactobacillus with lipolytic activity from the vinasse and their preliminary potential using as probiotics. AMB Expr 10, 91. https://doi.org/10.1186/s13568-020-01026-2.
Habib, S., Ahmad, S. A., Wan Johari, W. L., et al. (2020). Production of Lipopeptide Biosurfactant by a Hydrocarbon-Degrading Antarctic Rhodococcus. International journal of molecular sciences, 21(17), 6138. https://doi.org/10.3390/ijms21176138
Hou, J., Liu, W., Hu, W et al. (2021). Isolation, production and optimization of endogenous alkaline protease from in-situ sludge and its evaluation as sludge hydrolysis enhancer. Water Science and Technology, 83(11), pp.2700-2713.doi: https://doi.org/10.2166/wst.2021.167
Kumar, A., Mukhia, S., Kumar, N., et al. (2020). A broad temperature active lipase purified from a psychrotrophic bacterium of sikkim himalaya with potential application in detergent formulation. Frontiers in bioengineering and biotechnology, 8, p.642.
Lawniczak, L., Wozniak-Karczewska, M., Loibner, A. P., et al. (2020). Microbial Degradation of Hydrocarbons-Basic Principles for Bioremediation: A Review. Molecules (Basel, Switzerland), 25(4), 856. https://doi.org/10.3390/molecules25040856
Ma, M., Zheng, L., Yin, X. et al. (2021). Reconstruction and evaluation of oil-degrading consortia isolated from sediments of hydrothermal vents in the South Mid-Atlantic Ridge. Sci Rep 11, 1456. https://doi.org/10.1038/s41598-021-80991-5.
Mishra, P., Singh, U., Pandey, C.M., Mishra, P. and Pandey, G. (2019). Application of student’s t-test, analysis of variance, and covariance. Ann Card Anaesth; 22:407-11
Modi, V., and Dunbrack, R.L. (2019). A Structurally-Validated Multiple Sequence Alignment of 497 Human Protein Kinase Domains. Sci Rep 9, 19790. https://doi.org/10.1038/s41598-019-56499-4.
Mujumdar, S., Joshi, P. and Karve, N. (2019). Production, characterization, and applications of bioemulsifiers (BE) and biosurfactants (BS) produced by Acinetobacter spp.: A review. J Basic Microbiol. Mar;59(3):277-287. doi: 10.1002/jobm.201800364.
Patel, A.B., Shaikh, S., Jain, K.R., et al. (2020). Polycyclic Aromatic Hydrocarbons: Sources, Toxicity, and Remediation Approaches. Front. Microbiol. 11:562813. doi: 10.3389/fmicb.2020.562813
Rani, M., Weadge, J.T. and Jabaji, S. (2020). Isolation and Characterization of Biosurfactant-Producing Bacteria From Oil Well Batteries With Antimicrobial Activities Against Food-Borne and Plant Pathogens. Front. Microbiol. 11:64. doi: 10.3389/fmicb.2020.00064
Salwoom, L., Raja Abd Rahman, R.N.Z., Salleh, A.B., et al. (2019). Isolation, characterisation and lipase production of a cold-adapted bacterial strain isolated from Signy Island, Antarctica. Molecules, 24(4), doi:10.3390/molecules24040715.
Sani, F., Mokhtar, N.F., Mohamad Ali, et al. (2021). Enhanced Performance of Immobilized Rhizopus oryzae Lipase on Coated Porous Polypropylene Support with Additives. Catalysts, 11(3), p.303.
Silva, S.D., Gonçalves, I., Gomes de Almeida, F.C., et al. (2020). Soil bioremediation: Overview of technologies and trends. Energies, 13(18), p.4664.
Tekin, Z.H., Avci, E., Karasu, S. et al. (2020). Rapid determination of emulsion stability by rheology-based thermal loop test. LWT, 122, p.109037.


