Department of Biotechnology, Pramukh Swami Science and H.D. Arts Patel College, Kadi, Gujrat India
Article Publishing History
Received: 23/03/2019
Accepted After Revision: 30/05/2019
Increase in the need of petroleum results a remarkable rise in its prices. Therefore the need arises to discover alternative cheaper sources for fulfillment of world population demand. Hence the main objective to satisfy this need leads to the development of easier techniques using cheaper and eco-friendly raw materials for the production of Bio-ethanol. One such technique is production of Bio-Ethanol, an alternative to the present petrol and diesel, from fruit wastes by using well known yeast Saccharomyces cerevisiae RK1. The use of mixture of three fruits namely Banana, Grapes and Mango was used as a possible substrate for production of cellulosic ethanol by modifying parameters such as aeration. Pretreatment, hydrolysis and fermentation were carried out during this study. After fermentation of mixed fruits pulp without sucrose and fruits pulp with sucrose produced 0.67% ethanol and 1.32% ethanol respectively. Thus, the main objective behind this study is to produce bio-ethanol using cheaper substrates. The present study indicates a promising future for generation of ethanol at commercial scale from cellulosic wastes. Phylogenetic tree shows similarity with yeast sp.
Bioethanol, Eco-friendly, Fermentation, Fruit pulp, Saccharomyces Cerevisiae
Shah K. R, Vyas R, Patel G. Bioethanol Production from Pulp of Fruits. Oryzae.Biosc.Biotech.Res.Comm. 2019;12(2).
Shah K. R, Vyas R, Patel G. Bioethanol Production from Pulp of Fruits. Oryzae. Biosc.Biotech.Res.Comm. 2019;12(2). Available from: https://bit.ly/2ZFyRFT
Copyright © Shah et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
The fruits and vegetables are more prone to spoilage than cereals due to their nature and composition and this spoilage occurs at the time of harvestings, trasportation, storage, marketing and processing resulting in generation of wastes. Fruit wastes rich in reducing sugars are interesting feed stocks for production of first generation bio-ethanol (Joshi and Sandhu, 1996). Vegetable wastes are rich in cellulose, hemicellulose and lignin. So the production of second generation bio-ethanol from these could be an useful process (Zheng et al., 2009; Del Campo et al., 2006), earlier technologies have used reused fruits and vegetable waste to exploit the properties of food matrices to integrate human and animal diets, (Laufenberg et al., 2003).
Municipal solid waste has become a severe problem for disposal in developed and developing countries due to the shrinking landfill capacity. Biofuel is one of the best alternative fuels in order to trounce the energy crisis and solve pollution problems (Zainab et al., 2014).Since the need of bioethanol as a universal energy source has been increasing, the production of bioethanol must be increased using cheaper and eco-friendly raw materials. Bioethanol production from renewable ligno-cellulosic materials carries potential to reduce burgeoning world dependence on petroleum because of decreasing net emissions of carbon dioxide, the principal greenhouse gas, (James, 1997). According to Indian Agriculture Research Data Book 2004, the losses in fruits and vegetables are to the tunes of 30 percent. Taking estimated production of fruits and vegetables in India at 150 millions tones, the total waste generated comes to 50 million tons per annum. Efficient management of these wastes can help in preserving vital nutrients of our foods and feeds and bringing down the cost of production of processed foods, besides minimizing pollution hazards (Gautham and Gularia, 2007).
Due to energy crisis in 1970, it lead to the development of low – cost, sustainable and renewable energy resources such as ethanol.Ethanol has been described as one of the most exotic synthetic oxygen – containing organic chemicals because of its unique combination of properties as a solvent, a germicide, an antifreeze, a fuel, a depressant and especially of its versatility as a chemical intermediate for other organic chemicals.Investigations have carried out in Production of ethanol from food and agricultural waste now a day by bio-processing in research. Fruits Wastes such as banana, orange, pineapple and pea peels were saccharification and fermentation by co-culture of Aspergillus niger and Saccharomyces cerevisiae (Girisha Malhotra, 2013).Pretreated Banana and Mango fruit wastes hydrolyzed and fermented for ethanol Production (Arumugam et al., 2011).Other fruits wastes Grape, apple and banana, papaya or mixed fruit wastes using Saccharomyces cerevisiae (Kowshik, 2018).
The Carica papaya (pawpaw) agricultural waste, using dried active baker’syeast strain (Sacchromyces cerevisiae) also help in production of ethanol (Akin- Osannaiyeet al., 2008),Ethanol obtain from Different Fruit Wastes Using Saccharomyces cerevisiae (Janani et al., 2013) Hence bioethanol production could be the route to the effective utilization of the Municipal and agricultural waste (Shah et al., 2017).
In Fermentation of fruits by the action of yeast (Sacchromyces cerevisiae) that provide zymase enzyme result in conversion of sugar to produce alcohol, carbon dioxide and other by- products. The percentage yield of the alcohol production influenced by Nitrogenous compounds which are generally essential for the growth and development of yeast (Sacchromyces cerevisiae) in the fermentation processes (Nzelibe et al., 2001).The main aim of this research work is the biomasses used in bioethanol production are fruits pulp.The three biomasses with potential interest for bioethanol production have been used. The biomasses studied include grapes, banana and mango. The ripen fruit pulp raw materials fermented and enzymatic hydrolysis using yeast (Sacchromyces cerevisiae) have the ability to be converted Sugar into bioethanol. Basic objective of present studies:1.Isolation of microorganisms from fruit and vegetable wastes and characterization for saccharifying activity.2.Optimization of growth conditions for the fermentation process.3. Selection of efficient bacterial and yeast strains for alcohol production.
Materials and Methods
Isolation and Identification of Yeast
Yeast was isolated from fruits wastes by using GYE agar plate. Morphological character was studied by microscopy which shows oval and budding cell. Identification of yeast isolates was done by studying their morphological characteristics with the reference yeast, Saccharomyces cerevisiae cell identification was done by 18s rRNA sequencing method.
18s rRNA sequencing method
DNA Extraction
Lysis/homogenization: Cells grown in monolayer were lysed by suspending1-3 colonies aseptically and was mixed with 450 μl of “B Cube” lysis buffer in a 2 ml micro centrifuge tube and lyse the cells by repeated pipetting. 4μl of RNAse A and 250 μl of “B Cube” neutralization buffer were added. The content was vortex and incubated the tubes for 30 minutes at 65˚C in water bath. To minimize shearing of the DNA molecules, DNA solutions were mixed by inversion. Centrifuged the tubes for 20 minutes at 14,000rpm at 10˚C. Following centrifugation, the resulting viscous supernatant was transferred into a fresh 2 ml micro centrifuge tube without disturbing the pellet. 600 μl of “B Cube” binding buffer was added to the content and mixed thoroughly by pipetting and incubated the content at room temperature for 5 minutes. 600 μl of the contents was Transferred to a spin column placed in 2 ml collection tube. Centrifuged for 2 minutes at 14,000 rpm and discarded flow-through. The spin column and the collection tube were reassembled then transferred the remaining 600 μl of the lysate. Centrifuged for 2 minutes at 14,000rpm and discard flow-through. 500 μL “B Cube” washing buffer I was added to the spin column. Centrifuged at 14,000 rpm for 2 mins and discarded flow-through the spin column was Reassembled and added 500 μl “B Cube” washing buffer II and Centrifugated at 14,000 rpm for 2 mins and discarded flow-through. The spin column was transferred to a sterile 1.5-ml micro centrifuge tube. 100 μl of “B Cube” Elution buffer was added at the middle of spin column. Care should be taken to avoid touch with the filter. The tubes were incubated for 5 minutes at room temperature and Centrifuged at 6000 rpm for 1 min. The above mentioned step 14 and 15 were repeated for complete elution. The buffer in the micro centrifuge tube contains the DNA. DNA concentrations were measured by running aliquots on 1% agarose gel. The DNA samples were stored at -20°C until further use.
PCR Protocol
Polymerase Chain Reaction (PCR) is a process that uses primers to amplify specific cloned or genomic DNA sequences with the help of a very unique enzyme. PCR uses the enzyme DNA polymerase that directs the synthesis of DNA from deoxynucleotide substrates on a single-stranded DNA template. DNA polymerase adds nucleotides to the 3` end of a custom-designed oligonucleotide when it is annealed to a longer template DNA. Thus, if a synthetic oligonucleotide is annealed to a single-stranded template that contains a region complementary to the oligonucleotide, DNA polymerase can use the oligonucleotide as a primer and elongate its 3` end to generate an extended region of double stranded DNA.
Composition of the Taq Master Mix
Taq DNA polymerase is supplied in 2X Taq buffer 0.4mM dNTPs, 3.2mM MgCl20 and 02% bromophenol blue
Primer Details
| Primer Name | Sequence Details | Number of Base |
| LR7 | 5′ TAC TAC CAC CAA GAT CT 3′ | 17 |
| LROR | 5′ ACC CGC TGA ACT TAA GC 3′ | 17 |
Add 5 μL of isolated DNA in 25 μL of PCR reaction solution (1.5 μL of Forward Primer and Reverse Primer, 5 μL of deionized water, and 12 μL of Taq Master Mix). Perform PCR using the following thermal cycling conditions:
Denaturation
The DNA template is heated to 94°C. This breaks the weak hydrogen bonds that hold DNA strands together in a helix, allowing the strands to separate creating single stranded DNA.
Annealing
The mixture is cooled to anywhere from 51°C. This allows the primers to bind (anneal) to their complementary sequence in the template DNA.
Extension
The reaction is then heated to 72° C, the optimal temperature for DNA polymerase to act. DNA polymerase extends the primers, adding nucleotides onto the primer in a sequential manner, using the target DNA as a template.
Purification of PCR Production
Removed unincorporated PCR primers and dNTPs from PCR products by using Montage PCR Clean up kit (Millipore).The PCR product was sequenced using the primers. Sequencing reactions were performed using a ABI PRISM® BigDyeTM Terminator Cycle Sequencing Kits with AmpliTaq® DNA polymerase (FS enzyme) (Applied Biosystems).
Sequencing protocol
Single-pass sequencing was performed on each template using below 16s rRNA universal primers. The fluorescent-labeled fragments were purified from the unincorporated terminators with an ethanol precipitation protocol. The samples were resuspended in distilled water and subjected to electrophoresis in an ABI 3730xl sequencer (Applied Biosystems).
Bioinformatics Protocol
The sequence was blast using NCBI blast similarity search tool. The phylogeny analysis of query sequence with the closely related sequence of blast results wasperformed followed by multiple sequence alignment. The program MUSCLE 3.7 was used for multiple alignments of sequences.The resulting aligned sequences were cured using the program G blocks 0.91b.This G blocks eliminates poorly aligned positions and divergent regions (removes alignment noise).Finally, the program PhyML 3.0 aLRT was used for phylogeny analysis and HKY85 as Substitution model. PhyML was shown to be at least as accurate as other existing phylogeny programs using simulated data, while being one order of magnitude faster. PhyML was shown to be at least as accurate as other existing phylogeny programs using simulated data, while being one order of magnitude faster. The program Tree Dyn 198.3 was used for tree rendering.
Bioethanol Production
Preparation of raw materials
The waste fruits of banana, mango and grapes fruit waste were taken from the local market and washed properly to separate out the soil particles, spoilage particles and other adhering particles. 200 grams of each fruit were weighed and mixed in mixer with 200 ml of distilled water (1:1 ratio). 500 ml of substrate was taken in a 1000 ml flask.
Pretreatment of raw material
0.5 g NaCl and 0.25 KH2PO4 was added to substrates- fruits pulp and mixed well. The fruit pulp was separated into two flasks 200 ml and 100 ml and autoclaved for 20 minutes at 1210C and 15lb pressure. In acid hdyrolysis, acid concentrations between 0 to 4% v/v were used, high concentrations yielded high solubilized material. Dilute acid pretreatment was chosen since it is the most preferred and widely used method (Balat et al., 2008). Phosphoric acid was used since after neutralization of hydrolysates with NaOH, a salt formed and can remain in the hydrolysates, as it is used by microorganisms. The amount of reducing sugars was estimated by dinitrosalicylic acid (DNSA) method (Miller, 1959).
Inoculation of yeast
A 24 hr grown inoculums of isolated yeast in GYE broth was added to each flask 20 ml was inoculated into the flasks of 200ml fruits pulp. Also, we have added sucrose to the flasks containing fruit pulp, 1% sucrose was added to 200ml flasks respectively.
Fermentation
Lab-scale batch fermentations were carried out on 250 ml of pretreatment mix fruit waste of liquefied substrate. Another pretreatment mix fruit waste of liquefied flask with sucrose. inoculums were inoculated with the yeast culture (1 × 106 cell/ml), in 1 liter flask, under static conditions, at 28◦C. Samples were taken at 24, 72, 120, and 168 h to quantify ethanol production and sugar estimation respectively.
The maximum sugars are converted into bioethanol. The reducing sugar utilization during fermentation was analyzed by DNS method (Shah et al., 2017) and the bioethanol production was analyzed by using Dichromate method (Byadgi et al., 2015).
Recovery of the product
After the 36 hours fermentation processes a little sum each one sample was taken and centrifuged. The supernatant was gathered and the volume of the alcohol was dictated by the Dichromate method. At that point whatever is left of the sample was distilled by using batch distillation unit to collect the ethanol from distinctive fruit wastes (Sanchez, 2007). The distillation unit comprised of three parts: a reboiler, condenser funnel and a distillate flask. The vapors began to climb into the still head and passed through the condenser channel. The constant dissemination of cold water around the condenser funnel supported in cooling the alcohol rich vapors back to liquid state. The condensed liquid collected in the distillate and iodine test used to check the ethanol presence. The qualitative estimation of the presence of hydroxyl group was confirmed by FTIR. The quantitative estimation of bioethanol production was analyzed by using Dichromate method.
Results and Discussion
Isolation of yeast
Yeast cell were isolated from fruit waste and identified by morphological and 18s RNA sequencing method Sacchromyces cerevisiae RK1.it was also reported by Nahvi et al. (2002) isolated yeasts from fruits source.
Fruit Pulp
Currently, researchers have great interests on the production of ethanol by using biomass. According to Janani et al., mixture of Fruit pulp was chosen because it consists of useful sugars and monomers of sugars that could be fermented to produce ethanol and found suitable to be used as alternative energy source. The distilled liquid was found to be ethanol.
In the hydrolysis process the cellulose present in the substrate is converted into ethanol naturally or using the cellulose degrading bacteria after the pre-treatment process. Fermentation of sugars released during hydrolysis of substrates which was carried out by yeast at pH 6 and 28°C temperature, to convert into bioethanol. Fermentation was carried out with fermented samples being collected every twenty four hours for analysis of reducing sugar by DNS method for substrate utilization and is shown in the graph below. The bioethanol production was analyzed by using Dichromate method (Byadgi et al., 2015). The sample was distilled in the distillation unit. The presence of hydroxyl group was confirmed by FTIR.
In FTIR result graphs band stretch 3331.34 and 3329.06 both indicate O-H function group present in fruit sample without sucrose and fruit sample with sucrose respectively.Gene Bank Accession number of Saccharomyces cerevisiae RK1 (Gene bank Assesion No.MK225573).
In this study after fermentation Process by Saccharomyces cerevisiae mixed fruit pulps without sucrose produced 0.67gm % ethanol show in fig 1. Whereas with sugar 1.32gm% ethanol show in fig 4. Alcohol production was comparatively low concentration in work carried out by Gosavi et al., (2017). Gosavi also reported that fermented sugar present in fruit waste also affect alcohol production so it was observed clearly that fruits pulp containing sucrose increase alcohol production. Highest Alcohol production observed after 4 day (92 hrs) in both with and without Sucrose sample. Isolated Saccharomyces cerevisiae have high efficiency in alcohol production using less sugar as reported by Janani et al. Sugar level steadily decreases day by day in fermented broths. Experiments P-value of control is 0.000133 whereas P-value of ethanol produced without sucrose and with sucrose are 0.991334, 0.995558 respectively.
Table 1: Alcohol and Sugar estimation from fruits pulp without Sucrose
| Incubation Day | Alcohol in Gram% | sugar in Gram% |
| Day0 | 0 | 1.7 |
| Day2 | 0.63 | 0.7 |
| Day4 | 0.67 | 0.6 |
| Day6 | 0. 66 | 0.5 |
| Day8 | 0. 51 | 0.4 |
Table 2: Alcohol and Sugar estimation from fruits pulp Sucrose
| Incubation Day | Alcohol in Gram% | sugar in Gram% |
| Day0 | 0 | 2.53 |
| Day2 | 1.05 | 1.5 |
| Day4 | 1.24 | 0.24 |
| Day6 | 1.32 | 0.18 |
| Day8 | 1.02 | 0.11 |
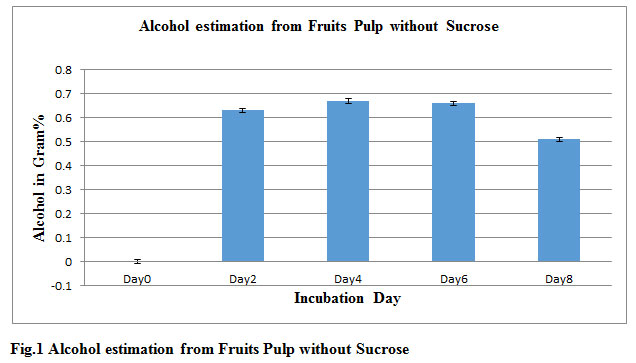 |
Figure 1: Alcohol estimation from Fruits Pulp without Sucrose |
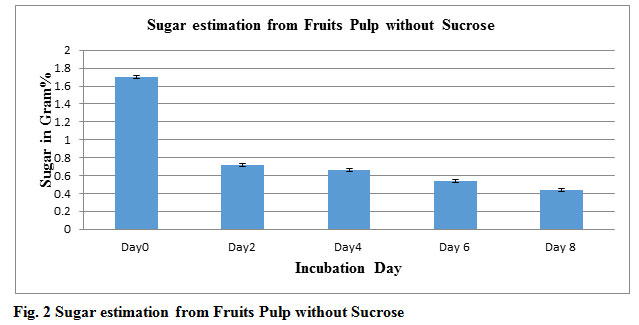 |
Figure 2: Sugar estimation from Fruits Pulp without Sucrose |
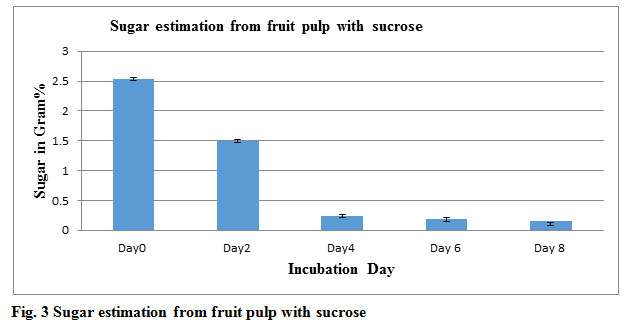 |
Figure 3: Sugar estimation from fruit pulp with sucrose |
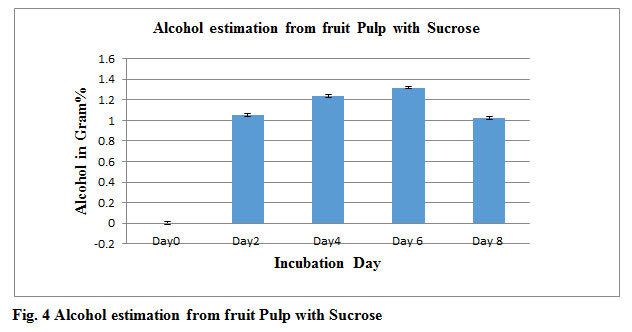 |
Figure 4: Alcohol estimation from fruit Pulp with Sucrose |
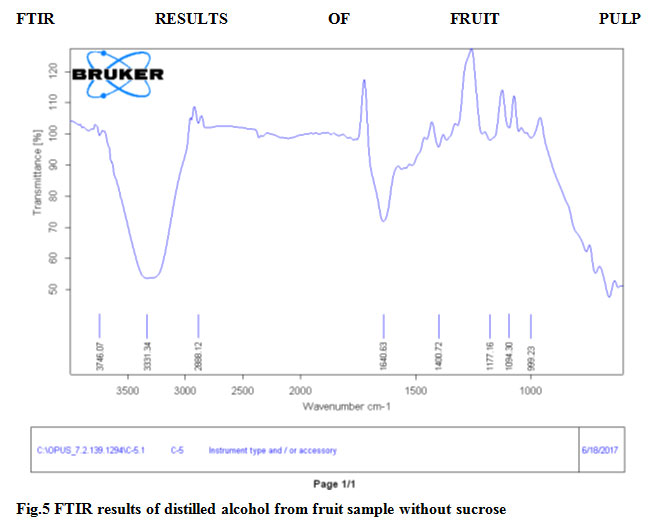 |
Figure 5: FTIR results of distilled alcohol from fruit sample without sucrose |
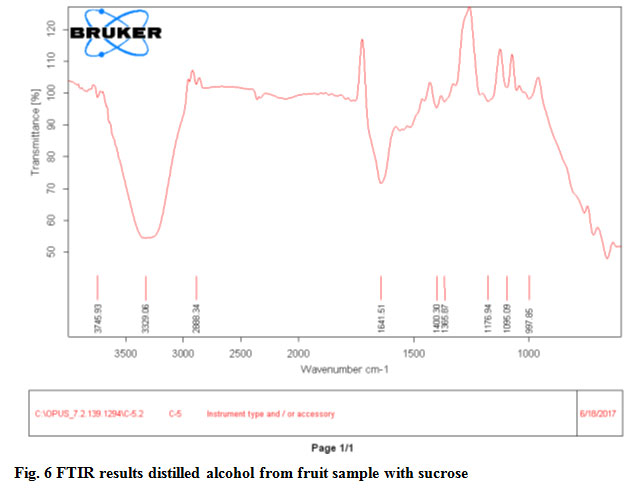 |
Figure 6: FTIR results distilled alcohol from fruit sample with sucrose |
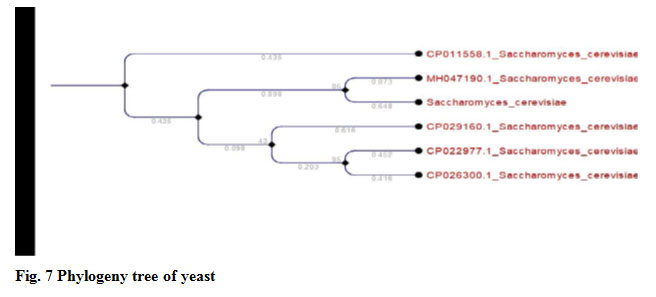 |
Figure 7: Phylogeny tree of yeast |
Conclusion
The present work deals with the studies on production of bioethanol from fruit pulp. Experiments were carried for Pretreatment of the substrate, Hydrolysis and Fermentation of the hydrolysate. The Saccharomyces cerevisiae RK1 was used to ferment the reducing sugar into ethanol. The presence of hydroxyl group was confirmed by FTIR. The ethanol yield as estimated using Dichromate method. Based on these results, it can be concluded that the production of bioethanol from fruit pulp was successful. We can predict that on re-distillation higher concentration of alcohol can be obtained, which can be used as biofuel. As this process is cost-effective and gives out no toxic substances, it could be produced at an industrial level.
Acknowledgements
The authors wish to express their thanks to the Department of Biotechnology, Pramukh Swami Science and H.D. Patel Arts College, Kadi.
Refrences
Akin- Osannaiye B. C., Nzelibe H. C. and Agbaji A. S. (2008): Ethanol production from Carica papaya (pawpaw) fruit waste. Asian Journal of Biochemistry, 3: 188-193.
Arumugam R., Manikandan M. (2011): Fermentation of Pretreated Hydrolyzates of Banana and Mango Fruit Wastes for Ethanol Production .biol. sci.; 2(2): 246-254.
Balat M.et al. (2008) : Progress in bioethanol processing., Progress in Energy and Combustion Science 34 551–573
Byadgi S. A. and Kalburgi P.B. (2015): Production of Bioethanol from Waste Paper. Procedia Environmental Sciences; 35: 555 – 562.
Castresana, J. (2000): Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540-552.
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM and Gascuel O (2008): Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res.1:36.
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. (1989): Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for16S ribosomal RNA. Nucleic Acids Res 17: 7843–7853.
Laufenberg, Benno Kunz Marianne Nystroem (2003):Transformation of vegetable waste into value added products:: (A) the upgrading concept; (B) practical implementations. Bioresource Technology. 87(2): 167-198.
Gosavi , Chaudhary Y. and Durve-Gupta A. (2017): Production of biofuel from fruits and vegetable wastes. European Journal of Biotechnology and Bioscience. 5(3): 69-73.
Hossain ABMS and Fazliny AR. (2010): Creation of alternative energy by bio- ethanol production from pineapple waste and the usage of its properties for engine African Journal of Microbiology. Research. 4(9):813-819.
del Campo I. Alegría M. Zazpe M. Echeverría (2006):Diluted acid hydrolysis pretreatment of agri-food wastes for bioethanol production.Industrial Crops and Products, 24(3): 214-221.
James D.McMillan(1997): Bio-ethanol production: Status and prospects. Renewable Energy (10): 295-302.
Janani K., Ketzi M., Megavathi S., Dr.Vinothkumar D., Dr. Ramesh Babu N.G.(2013): Comparative Studies of Ethanol Production from Different Fruit Wastes Using Saccharomyces cerevisiae. International Journal of Innovative Research in Science, Journal Engineering and Technology; 2(12): 2319-8753.
Joshi K. and Sandhu D.K. (1996):Preparation and evaluation of an animal feed byproduct produced by solid-state fermentation of apple pomace. Bioresource Technology; 56: 251-255.
Kowshik B. R. Vinay (2018): Comparative Studies of Ethanol Production from Different Fruit Wastes Using Saccharomyces cerevisiae [http://prezi. Com 28 January 2018]
Malhotra (2013): Alcohol Production from Fruit and Vegetable Waste. International Journal of Applied Engineering Research 8(15):1749-1756.
Matsakas L., Kekos D., Loizidou M. and Christakopoulos P. (2014): Utilization of household food waste for the production. Biotechnology for Biofuels; 7(4): 2-9.
Nahvi, I., Giti, E. and Lila, A., (2002): Isolation of a flocculating Saccharomyces cerevisiae and investigation of its performance in the fermentation of beet molasses to ethanol. Biomass and Bioenergy. 23: 481-486.
Sanchez, C., ( 2007): Lignocellulosic residues: biodegradation and bioconversion by fungi, Biotechnology Advanced journal, vol.27, (2), pp.185- 194.
Shah K., Sutaria D. and Parmar P. (2017): Bioethanol production from plant waste. Newest International Multidisciplinary Referred Journal; 3:30-35.
Talavera, G., and Castresana, J. (2007): Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology 56: 564-577.
Yi Zheng, Zhongli Pan, Ruihong Zhang (2009): Overview of biomass pretreatment for cellulosic ethanol production. Int J Agric & Biol Eng, 2(3): 51.
Zahid A., Muhammad G., Javaid A., Muhammad I. andAli S. (2011): Bioethanol productions from rice polish by optimization of dilute acid pretreatment and enzymatic hydrolysis. African Journal of Biotechnology; 11 (4): 992-998.
Zainab B. and Fakhra A. (2014): Production of Ethanol by fermentation process by using Yeast Saccharomyces cerevisae. International Research Journal of Environment Sciences Vol. 3(7): 24-32.


