1Department of Biological Sciences, Faculty of Sciences, King Abdulaziz University, Jeddah, KSA,
2Immunology Unit, King Fahad for Medical Research. King Abdulaziz University, Jeddah, KSA.
Corresponding author email: fbaseqab@kau.edu.sa
Article Publishing History
Received: 14/03/2022
Accepted After Revision: 29/05/2022
Hydrocarbons including phenol and toluene are considered as the major source of energy and raw material of different industrial products. Although toluene and phenol have various beneficial applications, many studies reported the massive negative impacts of these contaminants on the environment and on human health. This study aims to provide an assessment of biodegradation potential of Staphylococcus pasteuri isolated from the industrial area at the coast of Red Sea, Jeddah, Saudi Arabia.
From 29 isolates, a strain, that exhibits a notable growth on mineral salt medium supplemented with phenol and toluene as a sole carbon source, was chosen for further investigation. Different optimization conditions have been examined for optimal degradation; two concentrations of phenol and toluene (0.5% & 1.0%) and different incubation temperatures. Growth assessments was measured by optical density (OD) of phenol and toluene using spectrophotometer. Maximum OD for phenol and toluene: (ODmax= 0.787) and (ODmax= 0.969) compared to the abiotic control of (ODmax= 0.152) and (ODmax= 0.182) respectively. Degradation of phenol and toluene was also measured using high performance liquid chromatography (HPLC). In addition, molecular identification of the isolates was carried out using 16S rRNA analysis highlighted the isolated strain is Staphylococcus pasteuri (strain ATCC 51129) with the accession number (NR114435). This promising strain ATCC 51129 can be used in further biotechnological applications including oil biodegradation processes.
Biodegradation; HPLC; Hydrocarbon Contaminants; Staphylococcus
Basingab F. S, Nuqali W. E, Alaidaroos B. A, Alharbi N. M, Amasha R. H, Farsi R. M. Biodegradation Potential of Phenol and Toluene by Marine Staphylococcus pasteuri Isolated from the Red Sea, Saudi Arabia. Biosc.Biotech.Res.Comm. 2022;15(2).
Basingab F. S, Nuqali W. E, Alaidaroos B. A, Alharbi N. M, Amasha R. H, Farsi R. M. Biodegradation Potential of Phenol and Toluene by Marine Staphylococcus pasteuri Isolated from the Red Sea, Saudi Arabia. Biosc.Biotech.Res.Comm. 2022;15(2). Available from: <a href=”https://bit.ly/3z89hy3“>https://bit.ly/3z89hy3</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Phenol and toluene are highly toxic compounds compared to other types of hydrocarbons. Major problems associated with these compounds are their slow degradation rate which cause massive negative impacts on the environment. In addition, small concentrations of aromatic hydrocarbons such as benzene and toluene will cause vomiting, nausea, skin irritation, and inflammation. Constant exposure to these toxic components will cause long-last effects. In addition, these compounds can induce cancerous cells as a result of genetic mutations. All these contaminants and their processes cause harmful and lethal effects on the aquatic and marine organisms (Abdel-Shafy et al. 2016, Bianco et al. 2020, Ossai et al. 2020).
Various methods have been previously applied in an effort to overcome hydrocarbon contaminants such as adsorption or membrane filtration. Nevertheless, these methods are proven to be costly and inefficient especially when applied on large scales. Therefore, studies are shifting towards more cost-efficient and eco-friendly methods, one of which is biodegradation. Biodegradation is a principle where microorganisms are responsible for the partial or complete degradation or detoxification of contaminants. Many studies in recent years have focused on the degradation potential benefits of contaminants by some microorganisms i.e., bacteria, fungi, and algae (Basak et al. 2020, Giovanella et al. 2020, Khalel et al. 2021).
Importantly, some microorganisms can consume hydrocarbon contaminants and use them as an organic carbon source such as Aspergillus, Penicillium, and Bacillus (Varjani 2017, Hazim et al. 2019, Younis et al. 2020). However, application of bioremediation of hydrocarbons and petroleum at the contaminated sites is still not achieved on a large scale. This might be due to the poor biodiversity of microorganisms capable of degrading aliphatic and aromatic hydrocarbon compounds. Therefore, this study aims to isolate and identify a bacterium with capability of degrading phenol and toluene as hydrocarbon contaminants from aquatic contaminated sites in the Red Sea, Jeddah. In addition to the biochemical and molecular identification of the most efficient isolates with the optimal conditions affecting the process of biodegradation.
MATERIAL AND METHODS
The aromatic hydrocarbons: phenol and toluene were purchased from Sigma, with 98-99% purity. In addition, all other chemicals and reagents used in the study were obtained from Scharlau and Himedia.
Samples were collected from the industrial area in Red Sea, Jeddah at 21.4934694, 39. 17148778. 7 º. Samples were collected from aquatic regions and obtained at 10 – 30 cm depth. Five aquatic samples were placed into sterilized test cups in an aseptic condition. The collected samples were transported to the King Fahd Medical Research Centre (KFMRC), Microbiology laboratory using ice box and kept at refrigerator temperature 4oC until processed. Standard procedure of serial dilution was performed on all aquatic samples according to (Farsi et al. 2021). Diluted samples were mixed in the vortex for 10 minutes then inoculated into nutrient agar and incubated at 37ºC for 2-5 days. After the incubation period, each colony of mixed bacterial isolates were picked and sub-cultured three times on a new agar plate resulted in 29 pure isolates. One strain was chosen for further investigations.
The mineral salt medium (MSM) used for enrichment is composed of the following (g/l): KH₂PO4, 2.25; K2HPO4, 2.25; (NH4)SO4, 1.00; MgCl2·6H2O, 0.20; NaCl, 4.00; FeCl3 · 6H2O, 0.02 and CaCl2, 0.01 (pH 7.0). Bacterial isolates were grown on nutrient broth for 24 hrs. at 37ºC in rotating shaker. Then, samples were centrifuged at 14000 rpm for 10 minutes. After centrifuging, normal saline solution was added to isolates to dissolve completely. MSM was added as final step before inoculation. Isolate was inculcated in 50 ml of sterilized mineral salt medium (MSM) in Erlenmeyer flask which supplemented with different concentrations of 0.5%- and 1.0%-mM Phenol and Toluene as the only source of carbon for two weeks. Broth cultures were changed from Transparent white color to dark grey color. Bacterial growth was measured by spectrophotometer to evaluate optical density (OD) at 600 nm. Both negative control (abiotic MSM) and positive control (identified bacterium) were used in the study (Oberoi et al. 2015, Roccuzzo et al. 2020).
Estimation of biodegradation potential of phenol and toluene was carried out using Spectrophotometer and High-performance liquid chromatography (HPLC). For HPLC, 1 ml of each MSM samples was transmitted into spectrophotometer cuvette. Blank was the negative control. Growth measurements was estimated from inoculation (0 Time) to the 10th day. Absorbance of bacterial OD was measured at 600 nm.
Phenol and toluene degradation rate was estimated by HPLC (Shimadzu LCsolution Method). C18 column (4.6 μm,5-micron x150mm, Zorbax Eclipse plus) was utilized in stationary phase with wavelength 256 nm to detect phenol and toluene throughout biodegradation process. The degradation concentration was calculated based on the retention time and peak area (Oberoi et al. 2015, Xu et al. 2021). Different temperature degrees were applied during cultivation period to estimate the optimum temperature for the isolate. Cultivation the isolated bacteria were applied in 50 ml of sterilized MSM at two different temperatures (25 and 37℃) into shaking incubator for two weeks.
Two different concentrations (0.5 and 1.0 Mm) were supplemented in 50 ml of MSM, then incubated at 37℃ for two weeks. Evaluation the bacterial growth was obtained in different time intervals during the cultivation (Wongbunmak et al. 2021).
Biochemical tests, genomic DNA extraction of the bacterial isolate was done as according to the protocol by (Atashpaz et al. 2010). PCR amplification and 16 S rRNA gene sequencing was done using universal primers. The Forward primer 27F AGAGTTTGATCMTGGCTCAG and Reverse primer 1492 R TACGGYTACCTTGTTACGACTT) were used in the sequencing. Analysis of sequences was done by Blast search9 to find and compare between the similarities that found in NCBI GenBank. In addition, phylogenetic analysis was carried out for the blast results.
RESULTS AND DISCUSSION
This study was carried out to isolate bacteria capable of biodegrading phenol and toluene from the Red Sea coast, Jeddah, Saudi Arabia. Samples were collected from industrial aquatic regions. A total number of pure 29 isolates were found. The most active isolate, which grow well using phenol and toluene, was selected for identification. After characterization of bacteria, Staphylococcus pasteuri was chosen for further identification and estimating its potential for phenol and toluene biodegradation. Strain Staphylococcus pasteuri was Gram positive, cocci, colonies appearance was smooth and creamy with white color (Fig. 1).
In addition, Staphylococcus pasteuri was oxidase negative and gelatinase negative and catalase positive. In addition, antibiotic sensitivity test was done to evaluate resistance of the strain to some antibiotics (Table 1, Fig 2). Furthermore, change in color was noticed in MSM flasks during cultivation period from transparent white to dark grey (Hentati et al. 2021). Recent study for (Chaoui et al. 2022), referred that; the World Health Organization (WHO) has classified the strain of Staphylococcus maltophilia as a multidrug- resistant bacteria (MDR). Since the bacterium found to be related to a large range of infections.
Figure 1: Characteristics and biochemical analyses of isolated bacteria from Red Sea, Jeddah, Saudi Arabia. (A) Colony characteristics of isolated strain on nutrient agar plate. (B) Gram staining results of isolated bacteria showing violet cocci bacteria. (C) Negative result for oxidase production test. (D) Positive result for catalase production test.
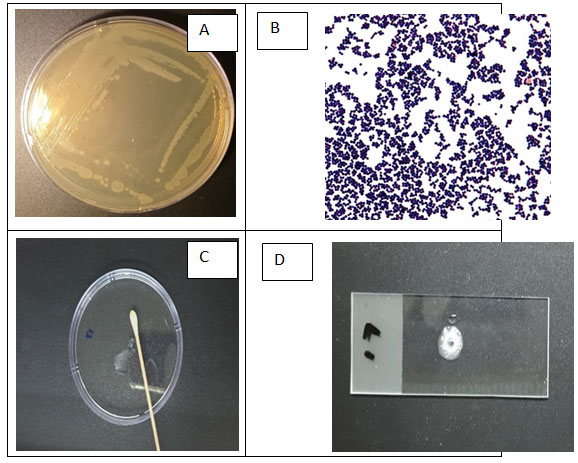
Figure 2: Antibiotic sensitivity test with different inhibition zone for the isolated Strain
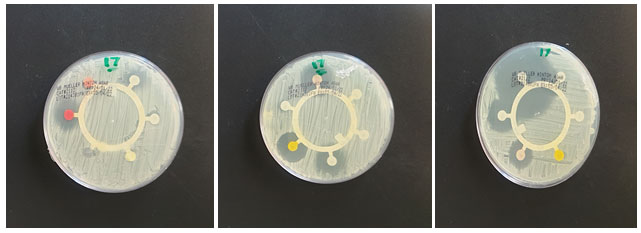
Table 1. Antibiotic sensitivity test for marine isolated strain
| Antibiotic | Results (mm) | Sensitivity |
| Ciprofloxacin | 22 | S |
| Amikacin | 16 | S |
| Imipenem | 46 | S |
| Ampicillin | – | R |
| Augmentin | – | R |
S: sensitive, I: intermediate, R: resistance.
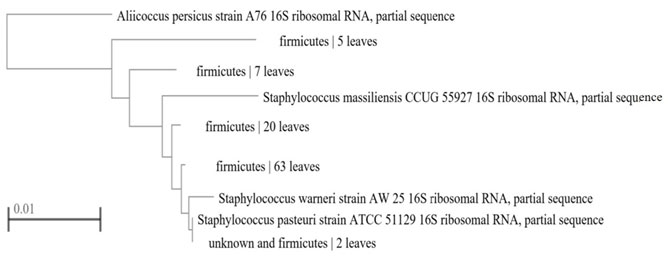
The molecular analysis of the strain exhibited a sequencing similarity with 99.85% to Staphylococcus pasteuri (strain ATCC 51129). The 16S rRNA gene sequencing (strain ATCC 51129) composed of 1419 nucleotides with accession number of NR114435 according to NCBI GenBank. Phylogenetic trait position of Staphylococcus pasteuri (strain ATCC 51129) is illustrated by a neighbor-joining dendrogram (Fig. 3).
Figure 3: Phylogenetic tree analyses based on 16S rRNA gene sequencing showing the position of Staphylococcus pasteuri strain ATCC 51129 with accession number NR114435.
Biodegradation of different concentrations of phenol and toluene was studied by Staphylococcus pasteuri. Staphylococcus pasteuri was cultivated in MSM medium supplemented with two different concentrations of phenol and toluene (0.5% and 1.0% mM). The best degradation rate was observed at 0.5 mM comparing to 1.0 mM. Measuring the growth of the strain illustrated by monitoring the OD at 600 nm for 10 days. The isolate showed a significant growth on phenol and toluene as (ODmax= 0.787) and (ODmax= 0.969) compared to the abiotic control of (ODmax= 0.152) and (ODmax= 0.182) respectively.
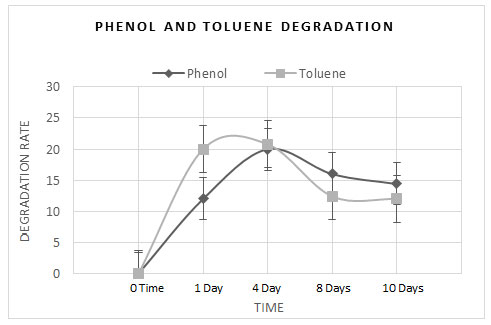
Moreover, HPLC analyses were achieved to evaluate the degradation rate of phenol and toluene at different interval times (0 time, 1 day, 4 days, 8 days and 10 days). Results showed a prominent capability of the strain to break down phenol and toluene as its sole carbon source. Figure 4 shows degradation percentages of phenol and toluene according to HPLC. The degradation rate of the isolate with phenol was as follows; 0% at 0 time, 12% at 1 day, 20% at 4 days, 16% at 8 days and 14.4% at 10 days.
The degradation rate for toluene was 0% at 0 time, 20% at 1 day, 20.8% at 4 days, 12.4% at 8 days and 12% at 10 days. The maximum degradation rate was observed at 4 days equivalents to 96 hours in both phenol and toluene. On the other hand, the least degradation rate was noticed after 10 days of monitoring. This might be cause of consuming most of the hydrocarbon compounds or due to the lethal effect of phenol and toluene on the isolate. Another study carried out using a bacterium isolated from soil contaminated areas (Staphylococcus pasteuri) has identified this bacterium as a degrader of poly-cyclic aromatic hydrocarbons in addition to n-alkane (Kiamarsi et al. 2019).
In addition, Imron et al. (2020) reported that; Staphylococcus sp. and Pseudomonas aeruginosa have proved a strong ability to perform biodegradation for diesel. These microorganisms were isolated from different environmental sites contaminated with diesel and considered them as hydrocarbons degrader. In addition, Staphylococcus pasteuri strain CO10 is another strain that considered as hydrocarbon degrader. Previous investigation on marine bacterium, Staphylococcus sp. CO100 strain has provide a great ability to biodegrade aliphatic hydrocarbons includes in crude oil with more than 70%. Growth of strain CO100 utilizing crude oil was evaluated by measuring OD using spectrophotometer. The maximum optical density was 0.65 compared to the optical density of control = 0.17, (Hentati et al. 2021).
Figure 4: Degradation rates of phenol and toluene by the strain ATCC 51129 obtained by HPLC
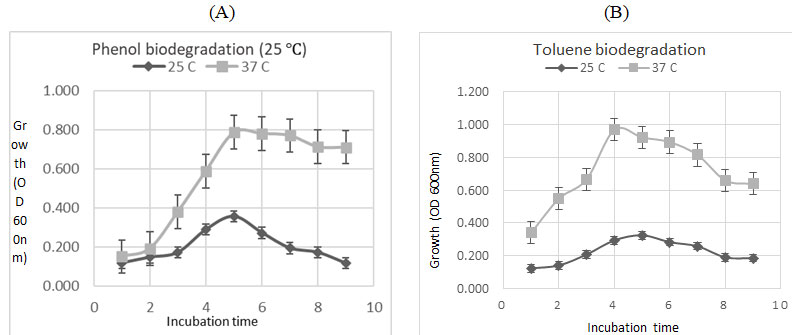
Investigation of different temperature degrees was carried out in the study. Estimation of biodegradation potential of phenol and toluene was done at 0.5mM with two different temperature (25 and 37℃) for ten days. At 37℃ temperatures, the maximum observation of OD was noticed after two days of incubation (OD= 0.787 and 0.969) for phenol and toluene respectively. On the other hand, OD at 25℃ was (0.357 and 0.325) for phenol and toluene (Fig. 5). The obtained result was in agreement with (Hentati et al. 2021) study on Staphylococcus sp. as a hydrocarbon degrader, which reported 37℃ as an optimum growth temperature.
The previous study investigated Staphylococcus sp. ability for biodegradation on a large temperature scale (15 to 55◦C). It should be noted, enzymatic activities also are one of the most important biological factors. The enzymatic activity showed significant inhibition with the increase of hydrocarbon contamination. Moreover, species of microorganisms is another important factor that will affect the success of biodegradation process. Staphylococcussp., Bacillus sp., Pseudomonas sp., and Acinetobacter sp. are well known to have high efficiency in degrading aliphatic and aromatic hydrocarbon compounds.
Furthermore, temperature variations affect hydrocarbon viscosity directly. High temperatures cause reducing to the viscosity and subsequently leads to better hydrocarbons diffusion as well as a better biodegradation process. Because the well-diffused contaminant is much easier to be degraded by the microorganisms. Generally, hydrocarbon’s viscosity decreases with low temperatures. Staphylococcus sp. is considered a promising bioagent and there is less research on Staphylococcus genus compared to the genera of Pseudomonas and Bacillus (Eddouaouda et al. 2012, Alrumman et al. 2015, Imron et al. 2020, Xu et al. 2021).
Figure 5: Effect of different incubation temperatures (25 and 37℃) on phenol and toluene biodegradation by the tested bacterium, (A): Biodegradation of phenol (B) Biodegradation of toluene.
CONCLUSION
Since the industrial revolution, hydrocarbons are becoming an integral part in daily life. Hence, massive amount of hydrocarbon waste is produced daily. In this study, several of bacterial strains isolated from aquatic contaminated sites. Among of other bacterial isolates evaluated for their biodegradation potential, Staphylococcus pasteuri was selected for further analyses as it exhibits biodegradation potential. Biochemical and molecular identification indicate that the isolate is Staphylococcus pasteuri with accession number (NR114435). Also, impact of different parameters was illustrated in the study such as temperatures and different phenol and toluene concentrations. The findings of the present study provided a handful information about the strain and its capability of phenol and toluene degradation which can be an excellent way of finding an alternative solution to achieve high biodegradation rates on a large scale.
Conflict of Interests: The author declares no conflicts of interests to disclose.
Funding: This research has been funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University under Grant no G: 1533-247-1440.
REFERENCES
Abdel-Shafy H I, and Mansour M S M (2016). A review on polycyclic aromatic hydrocarbons: Source, environmental impact, effect on human health and remediation. Egyptian Journal of Petroleum, 25(1), pp. 107–123.
Alrumman, S A, Standing D B ‘and’ Paton G I (2015). Effects of hydrocarbon contamination on soil microbial community and enzyme activity. Journal of King Saud University – Science. King Saud University, 27(1), pp. 31–41.
Atashpaz S, Khani S, Barzegari A et al. (2010). A robust universal method for extraction of genomic DNA from bacterial species. Microbiology, 79(4), 538-542.
Basak G, Hazra C ‘and; Sen R (2020). Biofunctionalized nanomaterials for in situ clean-up of hydrocarbon contamination. A quantum jumps in global bioremediation research. J. Environ. Manage. 256, 109913.
Bianco F, Race M, Papirio S at al. (2020). Removal of polycyclic aromatic hydrocarbons during anaerobic biostimulation of marine sediments. Sci. Total Environ. 709, 136141.
Chaoui, L. et al. (2022). Identification and assessment of antimicrobial resistance bacteria in a hemodialysis water treatment system. Journal of Water and Health, 20(2), pp. 441–449.
Eddouaouda K, Mnif S, Badis A, et al. (2012). Characterization of a novel biosurfactant produced by Staphylococcus sp. strain 1E with potential application on hydrocarbon bioremediation. J. Basic Microbiol. 52, 408-418.
Farsi, R. M. et al. (2021). Biodegradation of picric acid (2,4,6-trinitrophenol, TNP) by free and immobilized marine Enterococcus thailandicus isolated from the red sea, Saudi Arabia. Egyptian Journal of Aquatic Research. National Institute of Oceanography and Fisheries, 47(3), pp. 307–312.
Giovanella, P, Vieira G A, Otero I V R et al. (2020). Metal and organic pollutants bioremediation by extremophile microorganisms. J. Hazard. Mater. 382, 121024.
Hentati D, Cheffi M, Handrich F et al. (2021). Investigation of halotolerant marine Staphylococcus sp. CO100, as a promising hydrocarbon-degrading and biosurfactant-producing bacterium under saline conditions. Journal of Environmental Management. Elsevier Ltd, 277(June 2020), p. 111480.
Hazim R N ‘and’ Al-Ani M A (2019). Effect of petroleum hydrocarbons contamination on soil microorganisms and biodegradation. Rafidain J. Sci. 28(1), 13–22.
Imron M F, Kurniawan SB, Ismail N I et al. (2020). Future challenges in diesel biodegradation by bacteria isolates. a review. J. Clean. Prod. 251, 119716.
Kiamarsi Z, Soleimani M, Nezami A et al. (2019). Biodegradation of n-alkanes and polycyclic aromatic hydrocarbons using novel indigenous bacteria isolated from contaminated soils. Int. J. Environ. Sci. Technol. 16, 68056816.
Ossai I C, Ahmed A, Hassan A et al. (2020). Remediation of soil and water contaminated with petroleum hydrocarbon: A review. Environ. Technol. Innov. 17, 100526.
Roccuzzo S, Beckerman AP ‘and’ Trögl J (2020). New perspectives on the bioremediation of endocrine disrupting compounds from wastewater using algae, bacteria and fungi-based technologies. Int. J. Environ. Sci. Technol. 18, 89-106.
Wongbunmak, A. et al. (2021). A fixed-film bioscrubber of Microbacterium esteraromaticum SBS1-7 for toluene/styrene biodegradation. Journal of Hazardous Materials, 418, p. 126287.
Younis S A, Maitlo H A, Lee J et al. (2020). Nanotechnology-based sorption and membrane technologies for the treatment of petroleum-based pollutants in natural ecosystems and wastewater streams. Adv. Colloid Interface Sci. 275, 102071.


