1Laboratory of Microbiology and Experimental Medicine, Department of Zoology, University of Gour Banga, Malda, India
2Parasitology and Microbiology Research Laboratory, Department of Zoology, University of Burdwan, Burdwan, India
Corresponding author Email: samtropmed@gmail.com
Article Publishing History
Received: 12/01/2019
Accepted After Revision: 22/03/2019
The antibiotic resistances of bacterial isolates from various sources have been reported worldwide. The present study stands for the isolation and characterization of the bacteria isolated from the toys and clothes of children collected from children’s day care centre of Tarakeswar, Hooghly (WestBengal, India). Seven bacterial isolates (strain code: GUTY1, GUTY2, GUTY3, GUTY4, GUCL1, GUCL2 and GUCL3) were procured from the samples (toys and clothes) screened. Following conventional phenotypic and molecular characterization it was found that the children’s clothes and toys were contaminated with different potential pathogenic bacteria. The isolates GUTY1 and GUCL1 were identifi ed as E. coli, GUTY2 as Aneurinibacillus sp., GUCL3 and GUTY4 as Bacillus cereus where as GUTY3 as Lysinibacillus sp. and GUCL2 as Staphylococcus aureus. The disc diffusion susceptibility test revealed that the bacterial isolates had resistance to ampicillin only, or along with penicillin and/or rifampicin resistances.Thus, regular vigilance of antibiotic resistance of bacteria isolated from children’s toys and clothes is imperatively needed in order to combat the bacterial infection to the children especially in our part of the globe.
Toys And Clothes, Pathogenic Bacteria, 16s Rna Gene, Antibiotic Resistance, Mar Index
Bandyopadhyay R, Ghosh S, Chatterjee S, Mandal S. Bacteriological Profiling of Toys and Clothes of Children from Children’s Day Care Centre of Tarakeswar, Hooghly West Bengal, India. Biosc.Biotech.Res.Comm. 2019;12 (1).
Bandyopadhyay R, Ghosh S, Chatterjee S, Mandal S. Bacteriological Profiling of Toys and Clothes of Children from Children’s Day Care Centre of Tarakeswar, Hooghly West Bengal, India. Biosc.Biotech.Res.Comm. 2019;12(1). Available from: https://bit.ly/2XbVp0f
Copyright © Bandyopadhyay et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
Working family members have to rely on childcare centers for providing a safe, healthy and caring environment for their children. Children are very susceptible to contagious diseases as they have not been exposed to many microbial infections and also have no resistance to them, or sometimes have not received required immunizations (ARCH, 1996; Myo clinic, 2006). Children have high hand to mouth activity and obtain infections at an earlier age. Various infections have been documented in children in the childcare centers (Van et al, 1996). They play and eat close together without having proper hygiene habits.Children having diapers are more susceptible to the spread of diarrheal diseases due to poor or inadequate handwashing, diaper changing and environmental sanitation measures (Van et al, 1991). These contributing factors create infections in childcare centers. In children’s day care centre, children are commonly provided with toys and the safeties of these toys are usually not taken. Evidence has shown that belongings such as, toys and school-kits of children may easily be contaminated with microbes. Toys get passed from child to child and become contaminated through handling or by children putting their mouths to them, (Suviste, 1996, Agbulu et al 2015, Ali et al., 2016, Al-Easawi, et al., 2017 Johani et al., 2018).Children are very much susceptible to microbial diseases. Frequently they suffer from bacterial infections like pneumonia, urinary tract infections, diarrhoea, and so many. Most of these diseases occur due to poor hygiene practices. It was noticed that the children at the study areas had been suffering from various types of food-borne and water-borne diseases. There are approximately 7000 newborn deaths every day, amounting to 46% of all child deaths under the age of 5-years (WHO, 2016). The current study has been undertaken to determine bacterial contamination of children’s toys and clothes from West Bengal, India and to explore the bacterial antibiotic resistances.
Materials and Methods
Microbiological Analysis
Ten toys were collected from a children day care centre of Tarakeswar, Hooghly (West Bengal state, India) with which the children seemed to play regularly. Ten cloth samples (socks and shirts) of children were randomly selected for the microbiological examination. Toys were kept in sterile plastic bags. In the laboratory, 0.1% peptone broth (suffi cient to cover the toy) was poured into the bag and the toy was then rubbed from the outside of the bag to make sure that organisms were mixed with the broth. The broth was then shaken forcefully in the bag to make a uniform distribution of organisms, then serially diluted to 10-3 dilution and cultured following standard techniques (Maturin and Peeler, 2001). Ten clothes (socks and shirts) were selected for the microbiological examination. The area of clothes was swabbed using a sterile cotton swab, immersed in 0.1% peptone broth and used for culture.
Pour plating was done on nutrient agar for total bacterial count and on EMB agar for total coliform (TC) count. After pour plating Bacteria were isolated by sterile loop and streaked on nutrient agar plates and incubated at 30±1 oC in a BOD incubator for 24 h to obtain the isolated colonies. Pure culture was maintained on agar slants for further characterization and identification. Three replications were made for each sample tested. Average colony forming unit (CFU) of coliform and total bacteria count per milliliter of broth was recorded for a specific toy or specific cloth. Total bacterial count and TC count were categorized as ‘none detected’, ‘low’ (between no CFU and ≤103 CFU/mL), ‘moderate’ (>103 CFU/mL and or ‘high’ (<105 CFU/mL).
Determination of Phenotypic Identity of Isolated Bacteria
The shape, size, colour, margin and opacity of the isolated bacterial colonies were recorded. Phenotypic and biochemical characterization of the isolates were done following the standard protocol (Holt, 1984; Forbes et al., 2007). To study the bio-chemical properties, catalase, citrate utilization, nitrate reduction, indole production, methyl-red, Voges-Proskauer, urease, oxidase, and fermentation tests were done. For qualitative determination of enzymes, starch hydrolysis, lipase, protein hydrolysis, gelatin hydrolysis tests were done.
Molecular Characterization of Bacterial Isolates
The isolated bacteria from toys and clothes were identified both on the basis of 16S rRNA gene sequence analysis. Fingerprinting of the nucleotide was done following Lou and Golding (2007). The sequence data were aligned using the “ClustalW Submission Form” (http://www.ebi. ac.uk/clustalw) and analyzed by ClustalW (Thompson et al., 1994). Evolutionary distances were calculated using the method of Jukes and Cantor and the phylogenetic tree was prepared following the neighbor joining method (Saitou and Nei, 1987).
Bacterial Antibiotic Susceptibility and Mar Indices
The sensitivity of the bacterial isolates to antibiotics was tested following disc diffusion method (Bauer et al., 1966), as mentioned earlier by Mandal et al (2011) and Mandal et al (2016). The antibiotic discs (Hi-Media, India): ampicillin (10-μg/disc), chloramphenicol (30-μg/ disc), doxycycline (30-μg/disc), penicillin G (10 units/ disc), levofl oxacin (5 μg/ disc), nalidixic acid (30-μg/ disc), and rifampicin (5-μg/disc), were used. Antibiotic susceptibility test results, in terms of zone diameter of inhibition, were interpreted following the criteria of CLSI (2013). The MAR indices for the isolated bacteria were calculated following standard methods (Maloo et al, 2014; Krumperman et al., 1983).
Results and Discussion
The total bacteria count and TC count from the toys and clothes tested are shown in Table 1. The total bacteria count and TC count were categorized as ‘none detected’, ‘low’ (between no CFU and ≤103 CFU/mL), ‘moderate’ (>103 CFU/mL and or ‘high’ (<105 CFU/mL). Out of the 10 toys examined 3 were highly contaminated, 4 were moderately contaminated and the rest 3 had low contamination, while among 10 cloth samples tested, 2 were found highly contaminated, 3 were moderately contaminated and the rest 5 showed low bacterial contamination. A total of 7 bacterial isolates (strain code: GUTY1, GUTY2, GUTY3 and GUTY4 from toy samples, and GUCL1, GUCL2 and GUCL3 from cloth samples) were procured from children’s toys and clothes.The bacterial isolates GUTY1 and GUCL1 exhibited almost similar colony morphology and same biochemical properties: GUCL1 showed circular, creamy white and elevated colonies whereas GUTY1 formed spherical fl at whitish colony; both were rod shaped. The bacteria were gramnegative non-spore forming, and were positive for catalase, methyl red (MR), indole, nitrate reduction, starch and gelatin hydrolysis but negative for citrate utilization, oxidase, urease, coagulase and Vogues-Proskauer (VP) test. The bacterial isolates: GUTY2 and GUTY3 formed circular, white and elevated colonies and were gram-positive spore forming rod shaped. The GUTY2 strain was positive for catalase, MR reaction, but negative for VP reaction, indole production, citrate activity, nitrate reduction, H2S production, starch hydrolysis, gelatin hydrolysis, urease test, oxidase, coagulase test, lecithinase test, mannitol methyl red test.
The GUTY3 isolate was positive for catalase, indole, oxidase, nitrate reduction, starch and gelatin hydrolysis but negative for citrate utilization, methyl red and VP test. The bacterial isolate GUTY4 and GUCL3 showed more or less similar colony morphology. Both of them produced off-white, gummy and fl at colonies with entire margin. GUCL3 formed spherical colonies where as GUTY4 formed circular colonies. Two bacterial isolates, GUTY4 and GUCL3, were spore forming gram-positive rod, and were positive for catalase, MR reaction, citrate activity, nitrate reduction, H2S production, starch hydrolysis, gelatin hydrolysis, urease test, coagulase test, lecithinase test, and glucose-sucrose fermentation, but negative for VP reaction, indole production, oxidase and lactose fermentation. The GUCL2 isolate was non-spore forming gram-positive coccus, and positive for catalase test, oxidase test, H2S production and lipid hydrolysis but negative for MR test, indole, nitrate reduction, citrate utilization, VP and urease tests as well as starch and gelatin hydrolysis. The microorganisms produced acid and/or gas from glucose but did not utilize lactose and mannitol.
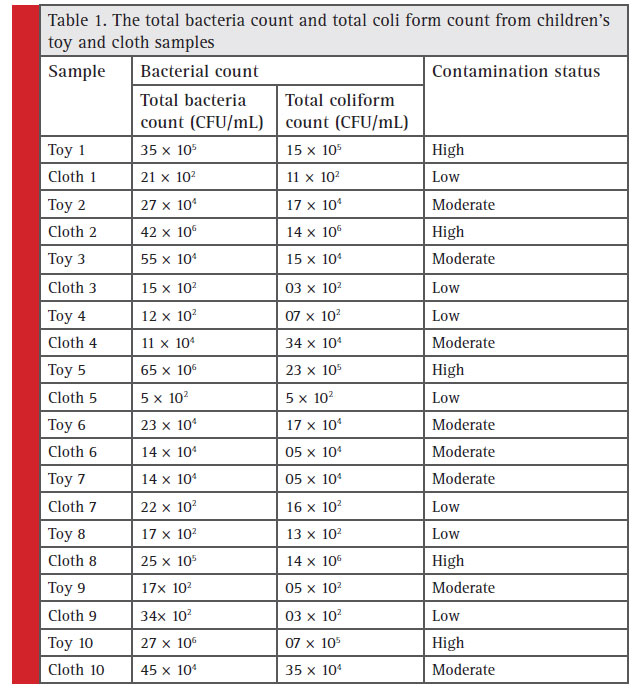 |
Table 1: The total bacteria count and total coli form count from children’s toy and cloth samples |
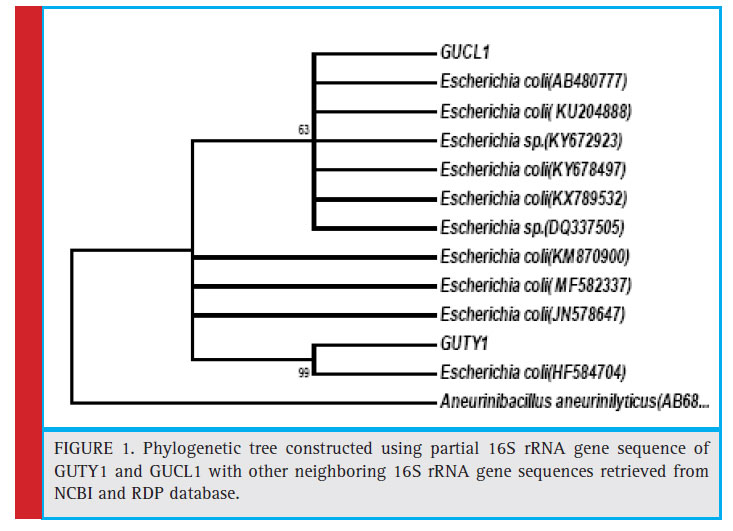 |
Figure 1: Phylogenetic tree constructed using partial 16S rRNA gene sequence of GUTY1 and GUCL1 with other neighboring 16S rRNA gene sequences retrieved from NCBI and RDP database. |
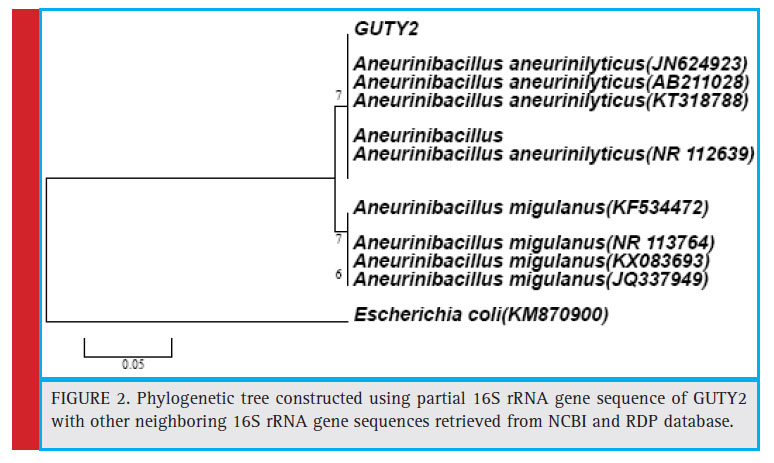 |
Figure 2: Phylogenetic tree constructed using partial 16S rRNA gene sequence of GUTY2 with other neighboring 16S rRNA gene sequences retrieved from NCBI and RDP database. |
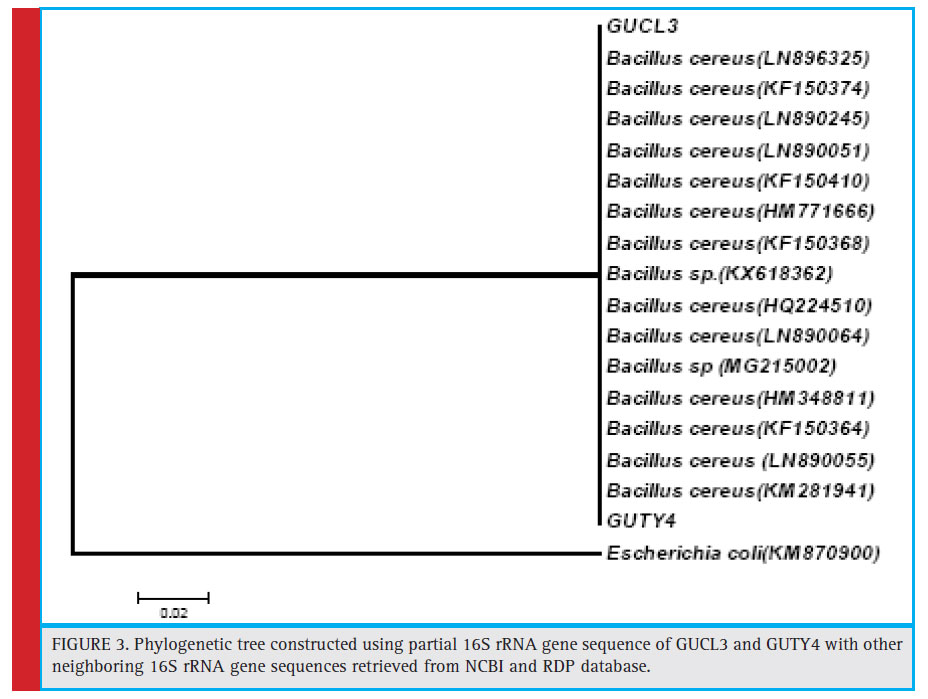 |
Figure 3: Phylogenetic tree constructed using partial 16S rRNA gene sequence of GUCL3 and GUTY4 with other neighboring 16S rRNA gene sequences retrieved from NCBI and RDP database |
KX789532, and DQ337505) with 63% bootstrap value and GUTY1 branched with E.coli (HF584704) with 99% bootstrap value (Figure 1).The GUTY2 isolate clustered with Aneurinibacillus aneurinilyticus (AB680012, JN624923, NR112639, AB211028, AB210964, and KT318788) and Aneurinibacillus migulanus (KF534472, NR113764, KX083693, and JQ337949),as represented in Figure 2.
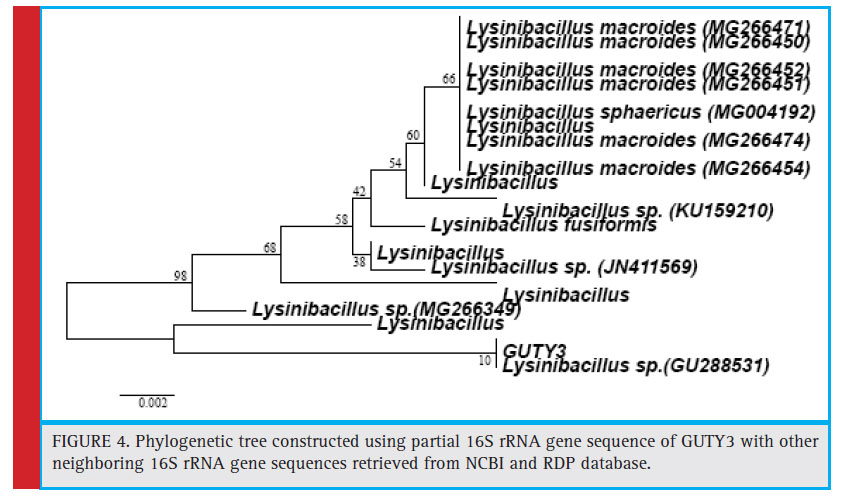 |
Figure 4: Phylogenetic tree constructed using partial 16S rRNA gene sequence of GUTY3 with other neighboring 16S rRNA gene sequences retrieved from NCBI and RDP database. |
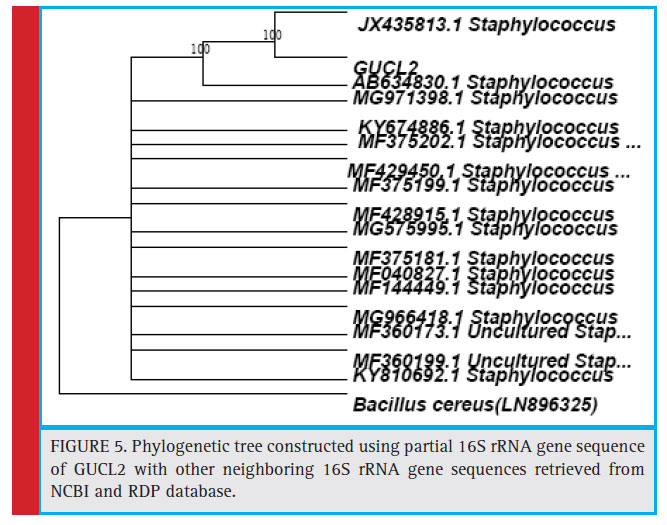 |
Figure 5: Phylogenetic tree constructed using partial 16S rRNA gene sequence of GUCL2 with other neighboring 16S rRNA gene sequences retrieved from NCBI and RDP database. |
The bacterial isolates GUCL3 and GUTY4 (Figure 3) were clustered with Bacillus cereus (LN890051, KM281941, HQ224510, KF150364, HM771666, LN890245, KF150374, KF150368, LN890064, HM348811, LN890055, KF150410, and LN896325). The GUTY3 isolate branched with Lysinibacillus sp. (GU288531) with 100% bootstrap value (Figure 4). The GUCL2 strains (Figure 4) has been clustered with Staphylococcus aureus (JX435813). Therefore, on the basis of phenotypic characteristics and phylogenetic tree analysis of bacterial partial 16S rRNA gene sequence, GUTY1 and GUCL1 were identifi ed as E. coli, GUTY2 as Aneurinibacillus sp., GUCL3 and GUTY4 both were identified as Bacillus cereus where as GUTY3 was identifi ed as Lysinibacillus sp. and GUCL2 as Staphylococcus aureus.
The isolates E.coli GUTY1 and E.coli GUCL1 showed sensitivity to chloramphenicol, doxycycline, penicillin G, levofl oxacin, nalidixic acid and rifampicin but resistant to ampicillin. The MAR index of E.coli (GUTY1 and GUCL1)isolates was 0.143. Aneurinibacillus sp. the GUTY2 isolate showed sensitivity to chloramphenicol, levofl oxacin,doxycycline, rifampicin,nalidixic acid but resistance to ampicillin and penicillin; the MAR index for the strain was 0.285.The Lysinibacillus sp. GUTY3 isolate and Bacillus cereus GUTY4 and GUCL3 isolates showed resistance to ampicillin with MAR index of 0.143. The Staphylococcus aureus GUCL2 isolate had resistance to ampicillin, penicillin G and rifampicin with MAR index of 0.428.
The bacterial isolates from the toys and clothes were identified on the basis of their phenotypic as well as molecular characters. Phylogenetic studies or culture independent techniques usually provide a better scope of predictable bacterial diversity in child-care environments (Kelley et al., 2004; McManus and Kelley, 2005). The 16SrRNA gene has been used in phylogenetic studies as it is evolutionary stable and highly conserved (Weisburg et al., 1991). Besides these highly conserved primer binding sites, the 16S rRNA gene sequences have hypervariable regions for species-specific signature sequences which are very important for the characterization and identification of bacterial strains. The 16S rDNA based identification of bacteria potentially offers a useful alternative when phenotypic characterization methods fail (Deancourt et al., 2000; Lee et al., 2002). It is scientific and objective method of identification of microorganisms (Tang et al., 1998). Present investigation revealed the presence of both spore-forming (B.cereus, Lysinibacillus sp., Aneurinibacillus sp.) and non-spore forming bacteria (E.coli and Staphylococcus aureus) from toys provided to the children and also from the shirts or socks belonged to the children of the day-care centres.
The isolated bacteria had resistance either to ampicillin (Bacillus cereus GUTY4 and GUCL3) or to both the antibiotics: ampicillin and penicillin G (E.coli GUTY1 and E. coli GUCL1). Current study also showed the presence of Staphylococcus aureus GUCL2 which had resistance to three antibiotics: ampicillin, penicillin G and rifampicin. The spore-forming bacterial isolates also had resistance to several antibiotics. Present study revealed the presence of gram positive, spore-forming Bacillus cereus GUCL3 and Bacillus cereus GUTY4 having the ability to resist adverse or dry conditions and could be able to grow at a variable range of temperatures (Meer et al. 1991; Von Stetten et al. 1998; Nicholson, 2002; Vilain et al, 2006). It has already been established as food-pathogens (Agata et al. 1995; Nester, 2004; Neely and Maley, 2000). The existence of Staphylococcus sp. was probably due to their colonization and survival on human skin and mucosa, without having any adverse effect (Kluytmans et al, 1997).
Similar bacterial diversity was reported by Lee et al. (2007). Microbial survival in clothes is determined by intrinsic factors including cloth properties or microbial features and also extrinsic factors comprising of environmental temperature, humidity etc. (Agbulu et al., 2010). Ivory and McCollum (1999) made an attempt to evaluate whether the presence of specifi c types of toys had any infl uence on the level of interactive play achieved by young children and concluded that cooperative play was signifi cantly more common in the presence of social toys. McCrea et al (2000) screened coagulase-negative Staphylococcus, Micrococcus sp, Bacillus sp, methicillin-resistant Staphylococcus aureus, Streptococcus and coliforms from the toys provided to children in neonatal intensive care unit (NICU) cots and reported that toys may be reservoirs for potential infantile nosocomial sepsis. Avila-Aguero (2004) isolated coagulase-negative Staphylococcus sp.; Bacillus spp; Staphylococcus aureus; alpha-hemolytic Streptococcus; Pseudomonas spp; Stenotrophomonas maltophilia, and other gram-negative organisms from the toys provided to the children in a children hospital and concluded that toys could be contaminated with hazardous bacteria and afforded unnecessary risks for nosocomial infection.
Merriman et al (2002) reported that hard toys had relatively low levels of contamination in comparison to soft toys. According to Stauber et al., (2013) faecal indicator bacterial levels on toys probably varied in respect to the water and sanitation conditions at home. Buttery et al. (1998) reported that nosocomial outbreak was associated with toys. The current study represents a take-home message that care should be taken for the cleaning and washing of socks and shirts/frocks of children on a regular basis. Toys in general provided to children in day-care centres pose an infectious risk and effective measures must be implemented to prevent the spread of infections via toys and other belongings, such as clothes.
Conclusion
From the present study it was found that the children’s clothes and toys were contaminated with several kinds of bacteria (Staphylococcus aureus, Bacillus cereus and E. coli) having the capacity to cause severe illnesses, and such kinds of children’s belonging might act as the vehicle of pathogenic bacteria in the community. From this point of view it is important to maintain proper hygiene of children. Parents should be more concern regarding this matter. The toys and clothes must be cleaned properly with disinfectants before everytime use. Overall, regular vigilance of drug resistant bacterial contaminants of children’s belongings is an important and imperative task in order to combat life-threatening infection to children caused with such microbial pathogens.
Author Contribution
Raktima Bandyopadhyay: performed experimental works and co-wrote the paper; Sucharita Ghosh: performed experimental works and co-wrote the paper; Soumendranath Chatterjee: characterized and analyzed the molecular identity of isolated bacteria; Shyamapada Mandal: designed the study and discussed the paper.
Conflicts of Interest
There was no confl ict of interest.
References
Al-Easawi, N.A.F. and Emran, F.K., (2017). A Microbial Survey of Second Hand Clothe Samples Collected from Baghdad Market. Journal of Al-Nahrain University-Science, 20(3), pp.127-136.
Ali, S., Al-Harbi, M.M. and Rahman, S.R., (2016) Bacterial Isolates, Present On Surface Of Toys In Child Care Centers, In Al- Rass City, Al-Qassim Reigon. KSA. ARCH National Resource Center for Respite and Crisis Care Services (1996). Preventing the spread of disease: Tips for
providers. Retrieved November 10, 2006. From http://www. archrespite.org/ archfs42.htm.
Agata, N., Ohta, M., Mori, M., & Isobe, M. (1995). A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS microbiology letters, 129(1), 17-19.
Agbulu, C. O., Gberikon, G. M., & Ajine, B. O. (2015). Isolation and characterization of microorganisms associated with second hand female undergarments and children wear sold in Makurdi Metropolis. International Journal of Current Microbiology and Applied Sciences, 4(1), 716-724.
Avila-Aguero, M. L., German, G., Paris, M. M., Herrera, J. F., & Safe Toys Study Group. (2004). Toys in a pediatric hospital: Are they a bacterial source?. American journal of infection control, 32(5), 287-290.
Bauer AW, Kirby WMM, Skerris JC, Turuck M. (1966)Antibiotic susceptibility testing by a standard single diffusion method. American Journal of Clinical Pathology 45: 494-496.
Blumenstyk, G., & McCollum, K. (1999). 2 Reports Question Utility and Accessibility in Distance Education. Chronicle of Higher Education, 45(32), A31-A31.
Buttery, J. P., Alabaster, S. J., Heine, R. G., Scott, S. M., Crutchfield, R. A., & Garland, S. M. (1998). Multiresistant Pseu domonas aeruginosa outbreak in a pediatric oncology ward related to bath toys. The Pediatric infectious disease journal, 17(6), 509-513.
Drancourt, M., Bollet, C., Carlioz, A., Martelin, R., Gayral, J. P., & Raoult, D. (2000). 16S ribosomal DNA sequence analysisof a large collection of environmental and clinical unidentifi -able bacterial isolates. Journal of clinical microbiology, 38(10), 3623-3630.
Francis, K. P., Mayr, R., von Stetten, F., Stewart, G. S., & Scherer, S. (1998). Discrimination of psychrotrophic and mesophilic strains of the Bacillus cereus group by PCR targeting of major cold shock protein genes. Applied and Environmental Microbiology, 64(9), 3525-3529.
Forbes BA, Sahm DF, Weissfeld AS. (2007) Bailey and Scott’s Diagnostic of Microbiology 2007; 12th Edition, Mosby (Elsevier), USA.
Henze, A S.V., Bockmühl, D. and Höfer, D., (2015.) Fabric-skin models to assess infection transfer for impetigo contagiosa in a kindergarten scenario. European Journal of Clinical Microbiology & Infectious Diseases, 34(6), pp.1153-1160.
Holt JG. Bergey’s manual of systematic bacteriology, Williams and Wilkins, Baltimore 1984.
Ibfelt, T., Engelund, E.H., Schultz, A.C. and Andersen, L.P., 2015. Effect of cleaning and disinfection of toys on infectious diseases and micro-organisms in daycare nurseries. Journal of Hospital Infection, 89(2), pp.109-115.
Johani, K., Abualsaud, D., Costa, D.M., Hu, H., Whiteley, G., Deva, A. and Vickery, K., (2018). Characterization of microbial community composition, antimicrobial resistance and biofi lm on intensive care surfaces. Journal of infection and public health, 11(3), pp. 418-424.
Jukes TH and Cantor CR. (1969). Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York. Academic Press.
Kelly, D., Campbell, J. I., King, T. P., Grant, G., Jansson, E. A., Coutts, A. G., … & Conway, S. (2004). Commensal anaerobic gut bacteria attenuate infl ammation by regulating nuclearcytoplasmic shuttling of PPAR- and RelA. Nature immunology, 5(1), 104.
Kluytmans, J., Van Belkum, A., & Verbrugh, H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clinical microbiology reviews, 10(3), 505-520.
Lee, P., Lee, S., Hong, S., & Chang, H. (2002). Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Applied microbiology and biotechnology, 58(5), 663-668.
Lee, M. K., Chiu, C. S., Chow, V. C., Lam, R. K., & Lai, R. W. (2007). Prevalence of hospital infection and antibiotic use at a university medical center in Hong Kong. Journal of Hospital Infection, 65(4), 341-347.
Lou M and Golding GB. (2007). Fingerprint: visual depiction of variation in multiple sequence alignments. Mol Ecol Notes, 7:908–914.
Mandal, MD., Mandal, S., and Pal, NK. Antibiotic resistance prevalence and pattern in environmental bacterial isolates. The Open Antimicrobial Agents Journal, 3, (2011), 45-52.
Mandal, S., Das, SN., Mandal, M., Plasmid mediated antibiotic and heavy metal co-resistance in bacterial isolates from Mahananda river water (Malda, India). Transl Med (Sunnyvale), 6, (2016) ,185. doi:10.4172/21611025.1000185
Maturin, L. J. (2001). Aerobic plate count. In: Bacteriological analytical manual online. http://www.cfsan.fda.gov/~ebam/ bam-3.html
McCrea, K. W., Hartford, O., Davis, S., Eidhin, D. N., Lina, G., Speziale, P., & Höök, M. (2000). The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology, 146(7), 1535-1546.
McManus, C. J., & Kelley, S. T. (2005). Molecular survey of aeroplane bacterial contamination. Journal of applied microbiology, 99(3), 502-508.
Meer, R. R., Baker, J., Bodyfelt, F. W., & Griffi ths, M. W. (1991). Psychrotrophic bacillus spp. in fl uid milk products: a review. Journal of food protection, 54(12), 969-979.
Merriman, E., Corwin, P., & Ikram, R. (2002). Toys are a potential source of cross-infection in general practitioners’ waiting rooms. Br J Gen Pract, 52(475), 138-140.
Mayo Clinic (2006). Children’s illness: Top 5 causes of missed school. Retrieved December 15. 2006. http://mayoclinic.com/health/childrens-conditions/CC00059
NCCLS. Performance standards for antimicrobial disk susceptibility tests. 7th Ed. Approved standard National Committee for Clinical Laboratory Standards document M2- A7, 20(2002),
NCCLS, Villanova, PA. Neely, A. N., & Maley, M. P. (2000). Survival of Enterococci and Staphylococci on hospital fabrics and plastic. Journal of
clinical microbiology, 38(2), 724-726.
Nester, Eugene, W., Anderson, DG., Roberts, CE., Pearsall, NN., (2004). Microbiology: A Human Perspective. McGraw-Hill Book Company, Inc., New York, U.S.A.
Nicholson, W. L. (2002). Roles of Bacillus endospores in the environment. Cellular and Molecular Life Sciences CMLS, 59(3), 410-416.
Saitou N and Nei M. (I987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol, 4: 406-425.
Stauber, C. E., Walters, A., de Aceituno, A. M. F., & Sobsey, M. D. (2013). Bacterial contamination on household toys and association with water, sanitation and hygiene conditions in Honduras. International journal of environmental research and public health, 10(4), 1586-1597.
Suviste J. (1996). The toy trap uncovered. Nursing Times. 92(10): 56-60. 3.
Tang, D., Dowell, E. H., & Virgin, L. N. (1998). Limit cycle behavior of an airfoil with a control surface. Journal of Fluids and Structures, 12(7), 839-858.
Thompson JD, Higgins DG, Gibson TJ. (1994). Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positionspecifi c gap penalties and weight matrix choice. Nucleic Acids Res, 22: 4673-4680.
Van R, Chuan-Chuan W, Morrow A, Pickering L. (1991). The effect of diaper type and overclothing on fecal contamination in day-care centers. JAMA. 265: 1840-1844.
Van R, Morrow A, Reves R, Pickering L. (1996). Environmental contamination in child day-care centers. Am J Epidemiol 1991; 133: 460-470.
Vilain, S., Luo, Y., Hildreth, M. B., & Brözel, V. S. (2006). Analysis of the life cycle of the soil saprophyte Bacillus cereus in liquid soil extract and in soil. Applied and Environmental Microbiology, 72(7), 4970-4977.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., & Lane, D. J. (1991). 16S ribosomal DNA amplifi cation for phylogenetic study. Journal of Bacteriology, 173(2), 697-703.


