Department of Bioinformatics, Maulana Azad National Institute of Technology, Bhopal, M.P. 452051, India
Article Publishing History
Received: 11/05/2016
Accepted After Revision: 05/06/2016
In the current development stages, the antimicrobial peptides (AMPs) acquired from animals and plants had revealed as a novel ameliorative procedure as comparison to the conventional antimicrobial approach. The plants provides one of the richest natural sources of AMPs; they use AMPs to govern their own defense system against various biotic and abiotic stresses. These peptides have shown the potential antimicrobial properties against pathogenic microbes. In this study, in silico attempt have been made with the availability of plant proteome sequence –Curcuma longa – commonly known as turmeric which have shown promising feature of being antimicrobial in nature. Therefore, using stringent computational tools, C. longa proteome sequences were processed, analyzed and carried out to look for new peptides with likely potential antimicrobial activity. Firstly, major C. longa proteins were digested in silico by means of the three well known protease enzymes. Then, selection of digested peptide were carried out on the basis of results of different kinds of multidimensional statistics analysis such as support vector machines (SVM), random forest (RF), artificial neural network (ANN) and discriminant analysis (DA). Finally, predicted digested peptides were further characterized to examine different physicochemical properties and then compared with the patent antimicrobial peptides available at CAMP database. This study reveals a novel 28 potential peptides which may be considered a likely potential anti-microbial peptide activity.
Antimicrobial Peptides, Artificial Neural Network,Discriminant Analysis, Non Antimicrobial Peptide, Randomforest, Support Vector Machine
Suryawanshi S. K, Chouhan U. Application of Bioinformatics in the Prediction and Identification of Potential Antimicrobial Peptides from Curcuma Longa. Biosc.Biotech.Res.Comm. 2016;9(2).
Suryawanshi S. K, Chouhan U. Application of Bioinformatics in the Prediction and Identification of Potential Antimicrobial Peptides from Curcuma Longa. Biosc.Biotech.Res.Comm. 2016;9(2). Available from: https://bit.ly/2pjZmnn
Introduction
Pathogenic invasions are very common due to continuous increasing in the microbes population worldwide and also the effective mechanism to combat with these infections are needed too. In past evolutionary era antibiotics have proven as strong microbicidal agents that ensure healthy survival of living organisms. Due to extensive uses, microorganisms started developing resistance against these antibiotics by changing its metabolic
pathways affected by these. In general, to kill microorganisms antibiotics target the cell wall and protein synthesis pathways. During resistance, bacteria modify its biological pathways that enable them escapes the attack of the antibiotics. Newer classes of antibiotics were generated and so as the resistant strains of bacteria, (Anaya 2013 and Mor 2015).
Consequently, the conventional antibiotics proved to be useless to a large extent against these multi-drug resistant strains. This threat resulted in the probe of newer antimicrobial agents, which could prove effective against these resistant strains and then came into picture the naturally occurring antimicrobial peptides (Andreu 1998). These amphiphilic peptides are present in closely every life form as Nature’s antibiotics. These peptides playrole as the first line of defense in case of invertebrates and as the components of innate immunity in case of vertebrates. These are either expressed constitutively or synthesized promptly upon induction (Akira 2001). Antimicrobial peptide is an alternative source of antibiotics was found earlier. AMP is produced by various microorganism such as fungi, bacteria, virus, Protista etc. It was found that plants also have the same properties to be effective against pathogenic microorganisms, (Krishnan 2016).
India have richest biodiversity in terms of flora and fauna. Different plants have medicinal properties to be effective against pathogenic microorganisms. Novel therapeutic modalities are needed due to occurrence of multiple drug resistant strains of pathogenic bacteria that lead to serious problem with respect to health perspectives. Mostly, antibiotics affect homeostasis of bacteria by interfering at specific site of action, which require few days for disabling. With these mechanism bacterial growth is inhibited but simultaneously selection pressure for resistant bacteria, (Smith 2007), Ortega‐Ramirez 2014 and Habiba 2016).
Besides, under such circumstances the bacteria morphology is normally preserved and a bacterium that is initially sensitive to the drug can develop resistance through mechanisms such as preventing the antibiotic from binding or entering the organism and producing an enzyme that inactivates the antibiotic or remodeling target molecules.Screening of Antimicrobial peptides (AMPs) from animals & plants are one of the hottest area of research to produce effective molecules as an additional substitutes to the currently used antimicrobial compound(Guralp 2013). In the innate immune system of the organism, these peptides act as effector molecules and represents first-line of defense against various pathogens. Unlike action of antibiotics, mostly AMPs follows mode of action by disrupting and permeating the microbial membrane), which is a mechanism that prevents a target organism from acquiring resistance to the peptide, (Zasloff 2002, Hancock 2015).
Curcuma longa L., commonly known as turmeric, is a spice used extensively in Eastern cuisine. The root of the plant is normally ground to yield a yellow colored powder, which then used for various applications. Some of the culinary uses of turmeric are as a coloring and flavoring agent and as a food preservative. Turmeric is also known to have medicinal applications dating back several millennia and has been an important component in the ancient Indian Ayurvedic medicines. Some of its documented modern medicinal uses include its wound healing capacity (Biswas 2003) and its use for the treatment of inflammation and tumors, (D’incalci 2005 Sahebrao 2014 and Zorofchian Moghadamtousi 2014).
In this present study we have provided an approach to find out some antimicrobial activity from its peptides using machine learning algorithms such as Artificial Neural Network, Discriminant Analysis, Random Forest and Support Vector Machine available at the interface of the CAMP database. Further, a comparison of physicochemical properties has been made between predicted AMPs and also with patent AMP available at CAMP database.
Material And Methods
In this study, we download 165 protein sequences from NCBI with the hit of keyword “Curcuma longa”. Enormous peptide will be generated using these protein sequences by in silico digestion which is further evaluate the performance of our methodology. The protein sequences of turmeric proteins were subjected to in-silico proteolysis using Expasy Server(Gasteiger 2005). Specific hydrolytic enzymes action were performed here to determine the release of fragmented peptides from precursor proteins. Availability of peptide cutter tool at expasy server, which have different enzymatic as well as chemical option to digest protein sequences to peptides. In our proposed work, we implemented in silico digestion with the enzymatic action only, randomly selected three enzymes i.e., trypsin, protein kinase and pepsin out of 38 available at expasy server.
Digested peptides were subjected to antimicrobial activity with the use of the Prediction Antimicrobial Peptides tool available in the CAMP database. Four different multivariate statistical methods were implemented for efficient prediction of AMP: Support Vector Machines (SVM), Random Forest (RF), Artificial Neural Network (ANN) and Discriminant Analysis (DA). Waghu et al has been described the detail of model development and evaluation. Except ANN, prediction score as a results are presented in the numeric form and peptides are classified as AMPs or Non-AMPs. We consider released peptides as AMP, in case when all the four statistical methods predicted as AMP (Waghu 2014). Further, these peptide characterize for its physicochemical properties.
The physicochemical properties of predicted AMPs were calculated with the use of available of free software and online algorithms. Protein length, peptide mass, hydrophobicity, steric hindrance, side bulk, hydropathicity, amphipathicity, hydrophilicity, net hydrogen, charge, pI, boman index, hydropathy index, aliphatic index, instability index, GRAVY (grand average of hydropathicity) are the physicochemical properties of predicted AMP were computed with the use of freely available tools AntiCP is web based prediction server for Anticancer peptides (Tyagi 2013) and APD2: Antimicrobial Peptide Calculator and Predictor(Wang 2009).
Results & Discussion
In Silico Proteolysis Of C. Longa Proteins
Different techniques are implemented to identify and produce significantly active peptides from different sources of proteins. Naturally, peptides present in the protein sequence which is released by enzymatic action or by the action of enzymes derived from different microorganisms. Mostly proteolytic enzymes such as trypsin, papain, pepsin, chymotrypsin and pancreatic elastase are used to digest proteins to peptides. Further, digested peptides should be refined such as fractionation, separation and purification followed by evaluation of antimicrobial activity. With advent of research and development, now a days the cleavage site of enzymes are known as well as properties of peptides, due to this it is possible to search new AMP with bioinformatics tools (Keil 1992). Whole strategy are based on the computer simulation of proteolysis and application of multivariate statistical methods to determine potentially antimicrobial peptides released from the analyzed proteins. So many tools available online for the proteolysis such as PMAP(Igarashi 2009)and Peptide Cutter(Keil 1986). In this study, 3 enzymes from the peptide cutter were used to simulate the proteolysis of C. longa proteins and produces thousands of different fragments. Prediction of antimicrobial activity were based on specific ranges of 6 to 30 amino acids. As a resultant only 28 peptides out of 1813 non redundant peptides fragments were further evaluated for antimicrobial properties.
Prediction Of Antimicrobial Activity Of Peptides Released During In Silico Proteolysis Of C. Longa Proteins.
Effective tools are available at different database for the prediction of antimicrobial activity of peptides such as APD (Wang 2004), Bactibase (Hammami 2010), and PhytAMP(Hammami 2009). In this study, prediction tools available in CAMP database were used which is based on various machine learning algorithms such as SVM, RF, ANN and DA. The results accuracy for different statistical models ranges from 87 to 93% and its prediction of antimicrobial activity is regardless of length. Firstly, the antimicrobial potential of C. longa protein fragments produced after proteolysis were evaluated with the use of four statistical models available in the CAMP database. The results were used to manually select peptides where a positive result was reported throughout all the four statistical models (Dziuba 2014). A total of 28 potentially antimicrobial peptides with the prediction scores higher than 0.45 for at least three statistical models were thus selected (Table–2).
Physicochemical Characteristics of Amps
Physicochemical properties of 28 predicted AMPs from C. longa were calculated and compared with the AMP patent database available at CAMP. Table 1 represent the comparison of amino acid frequency and amino acid composition (%) between 28 predicted AMPs and AMP patent database. Most frequent amino acids found in AMP are ten which covers more than 66% in the proteolytic cleavage of C. longa and 77% of overall amino acid composition i.e., G A V L I K N S T E in patent database. On the other hand, 44 & 23% of amino acid composition covers M P W Q Y C F D R H respectively. The content of amino acids in the proteolytic peptides was calculated and compared with the patent peptide database. The results indicates that frequency of certain amino acids are more common in the AMPs and can influence their biological activity (Bevilacqua 2015).
| Table 1. Comparison of Amino acid frequency and its composition between predicted AMPs from C. longa and patent AMPs available at CAMP | ||||
| AMPs in C. longa Proteins | AMPs Patent Database at CAMP | |||
| One Letter Code of Amino Acid | Amino Acid Frequency | Amino Acid Composition (%) | Amino Acid Frequency | Amino Acid Composition (%) |
| G | 37 | 8.02603 | 25096 | 7.20841 |
| A | 32 | 6.94143 | 24649 | 7.08001 |
| V | 25 | 5.42299 | 24067 | 6.91284 |
| L | 33 | 7.15835 | 41118 | 11.8105 |
| I | 70 | 15.1844 | 26000 | 7.46807 |
| M | 4 | 0.86768 | 7961 | 2.28666 |
| P | 29 | 6.29067 | 9544 | 2.74135 |
| F | 15 | 3.2538 | 15310 | 4.39754 |
| W | 7 | 1.51844 | 2302 | 0.66121 |
| N | 17 | 3.68764 | 17578 | 5.04899 |
| Q | 20 | 4.33839 | 13487 | 3.87392 |
| S | 28 | 6.07375 | 25356 | 7.28309 |
| T | 10 | 2.1692 | 17908 | 5.14377 |
| Y | 8 | 1.73536 | 10752 | 3.08833 |
| C | 16 | 3.47072 | 4884 | 1.40285 |
| D | 4 | 0.86768 | 15706 | 4.51129 |
| E | 11 | 2.38612 | 19571 | 5.62144 |
| K | 43 | 9.32755 | 25050 | 7.1952 |
| R | 38 | 8.24295 | 16227 | 4.66094 |
| H | 14 | 3.03688 | 5583 | 1.60362 |
| Total | 461 | 100 | 348149 | 100 |
The Majority of antimicrobial peptides are cationic amphipathic peptides, also containing one or more hydrophobic residues. Hydrophobic amino acids such as I G L accounts for more than 30 percent of analyzed AMPs from C. longa and its play important role in secondary structure formation and interactions with bacterial membrane(Taniguchi 2016). The cationic and hydrophobic characters peptides affect the antimicrobial activity. The analysis of amino acid content of antimicrobial peptides revealed many fragments with the predominance of one or several amino acids. In comparison with all AMPs listed in the patent database, amino acid such as V & S are less frequently encountered from C. longa. On average, the evaluated peptides in both case have higher content of I, K & G. In both the case, result shows significant frequency of serine to be antimicrobial in nature(Pushpanathan 2013).
As earlier discuss that 165 proteins from C. longa were processed and as a resultant only 28 peptides predicted to have antimicrobial activity as represented in table 2. Proteolysis of 165 proteins generates 5858 peptides fragments, after removal of redundancy 1813 peptides remain to be non-redundant. These peptides were predicted for AMP through various algorithms such as support vector machine (SVM), random forest (RF), artificial neural network (ANN) and discriminant classifier (DAC). Only 3 out of 4 algorithms provides accuracy in numeric form i.e. above 0.5 and ANN provides only information that whether peptides are AMP or NAMP, then peptide consider as Antimicrobial peptide (AMP). On the basis of prediction evaluation of these peptides only 28 peptides out of 1813 peptides found to be antimicrobial in nature. Due to diversity in these peptide sequences accuracy and physicochemical properties are also varies from peptide to peptide. Highest accuracy more than 90 percent among all the three algorithms were found in only one peptide sequences i.e., GAIIGNRKIKLQPHIIIRIID. More than 90 percent accuracy was found among two algorithms were found in nine peptides i.e., VWVKPWLHFK, VLGFPSKPIGLFIK, RKRHMY, KKPHRY, IIPKKSIY, GIVCSAVNSETGEQVAIKKIGNA, GGIRCPLTVVQSRGIGTIISSP, GAIIGDSKIRLQPHIIKRIIS,LGIQAFQPILVEGSAICLHPLVCK and other remaining 18 peptides have comparatively lower accuracy below 90 percent.
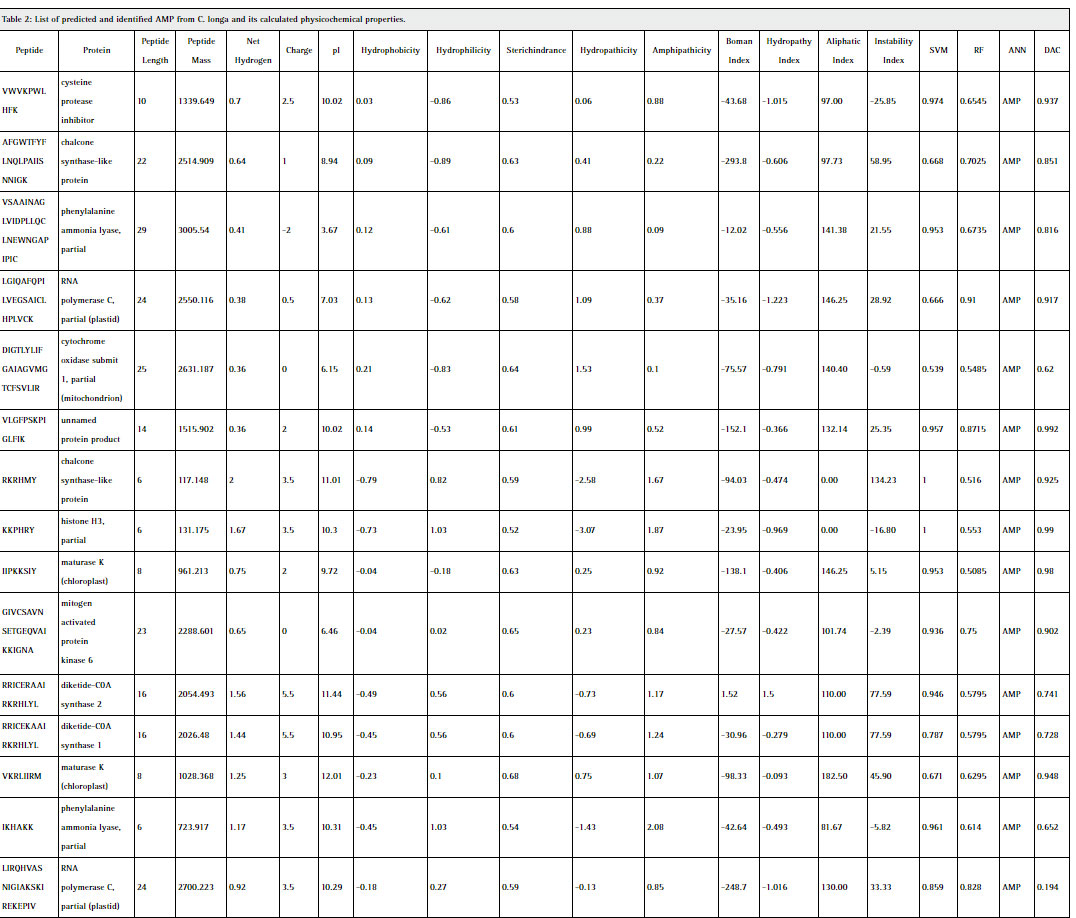 |
Table 2: List of predicted and identified AMP from C. longa and its calculated physicochemical properties. |
Statistical analysis of the physicochemical properties of 28 predicted AMP represented in table 3. Some valuable information inferred from these results in terms of significant correlation. (Chan 2003) Peptide length is significantly correlated with the peptide mass with 0.967, hydrophobicity with hydropathicity with 0.932, steric hinderance & steric bulk with peptide mass with 0.356, Hydropathicity with GRAVY with 0.999, Amphipathicity with hydrophilicity with 0.874, Net hydrogen with hydrophilicity with 0.842, charge with pI with 0.780, hydropathy index with aliphatic index with 0.777, instability index with steric hinderance with 0.349. Figure 1 shows graphical representation of various physicochemical properties of predicted AMP from C. longa.The selected peptides in our work are potentially antimicrobial AMPs have 6 to 30 amino acids, which corresponds to peptide mass of 117.175 Da to 3427.126 Da. The general classification of AMPs include cationic peptides which can be divided in three subclasses: linear peptides forming helical structures, cysteine-rich open-ended peptides containing single or several disulfide bridges and molecules rich in specific amino acids, such as proline, glycine, histidine and tryptophan. Majority of antimicrobial peptides with positive charges known as cationic antimicrobial peptides (CAPs). In our study, 85.71%, 3.57% & 7.14% peptides are found cationic, anionic and neutral charge in nature.
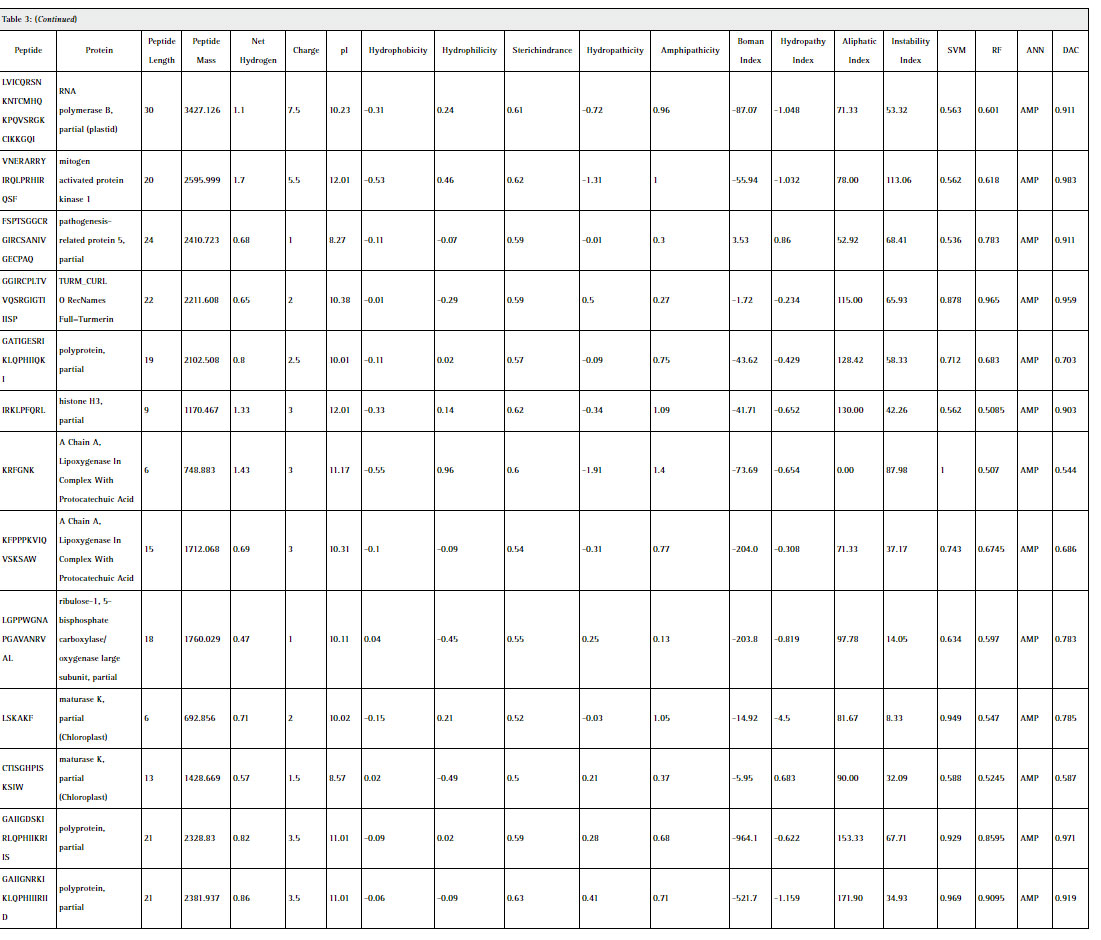 |
Table 3: (Continued) |
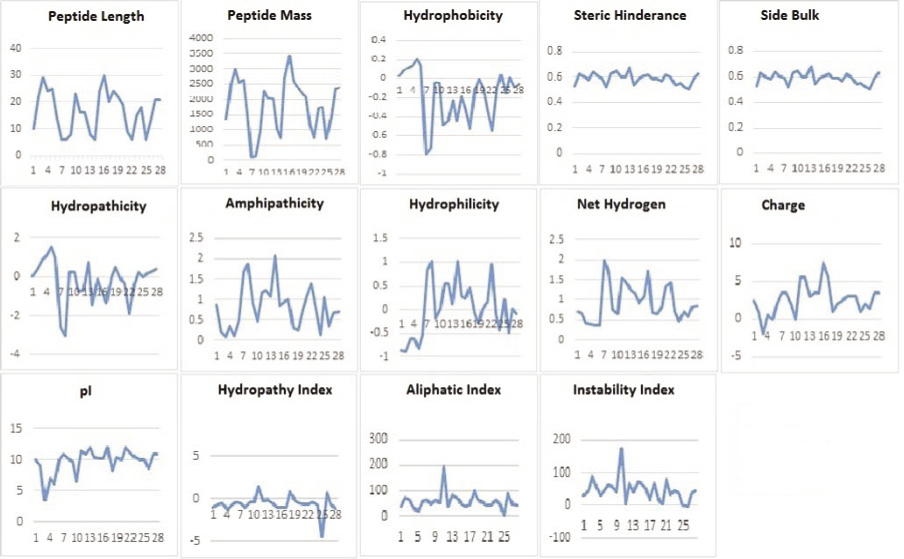 |
Figure 1: Graphical representation of different physicochemical properties of predicted AMPs |
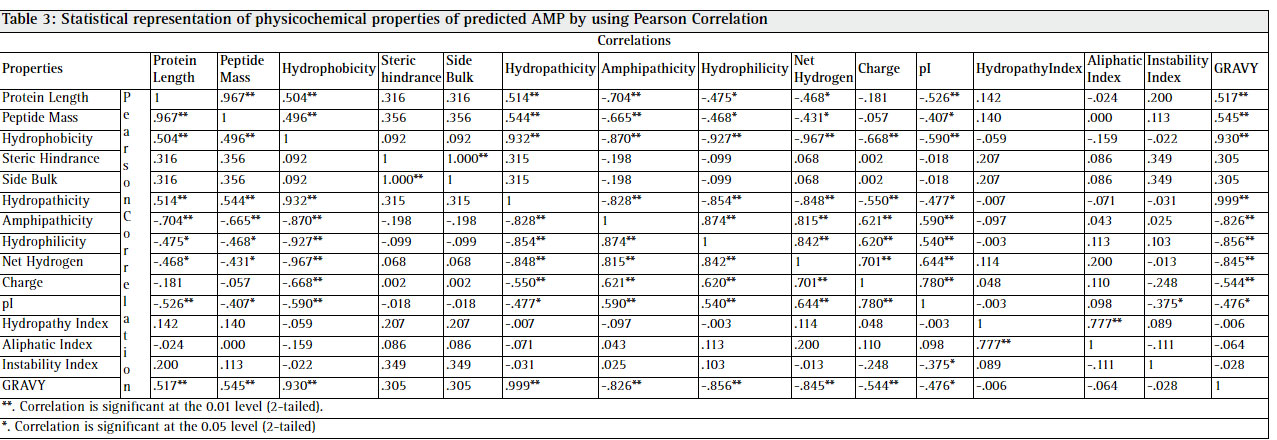 |
Table 3: Statistical representation of physicochemical properties of predicted AMP by using Pearson Correlation |
Conclusion
Peptide drug development is one of the recent approaches which have potential to replace the antibiotics and be effective in multidrug resistance organism. Here, in our work we concluded that 28 shorter fragment of turmeric protein have potential to be AMP. Synthetically designed these peptide will shows the promising feature to be effective against microorganism progression. Predicted and identified AMP from turmeric which will show positive result in wet lab experiments. Artificial synthesis of peptide requires efficient design, here we provide in silico analyzed curated peptides which needs further wet lab validation before practicing/applying them for therapeutics and industrial application.
Acknowledgments
The authors are highly thankful to the Ministry of Human Resource Development, Government of India and Department of Bioinformatics, MANIT, Bhopal, India for providing support in the form of Bioinformatics infrastructure facility to carry out this work.
References
Akira, S., Takeda, K., Kaisho, T. (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nature Immunology, Vol. 2, No. 8,pp. 675-80.
Anaya, L., López M., Ochoa Z., Alejandra (2013), Bacterial resistance to cationic antimicrobial peptides. Critical Reviews in Microbiology, Vol. 39, No. 2,pp. 180-95.
Andreu, D., Rivas, L. (1998) Animal antimicrobial peptides: an overview, Peptide Science, Vol. 47, No. 6,pp. 415-33.
Bevilacqua, M.P., Benitez, D., Deming, T.J., Hanson, J.A., Koziol, L. (2015) Composition with high antimicrobial activity and low toxicity. US Patent 20, 150,366,193.
Biswas, T.K., Mukherjee, B. (2003) Plant medicines of Indian origin for wound healing activity: a review. The International Journal of Lower Extremity Wounds, Vol. 2, No. 1, pp. 25-39.
D’Incalci, M., Steward, W.P,Gescher, A.J. (2005) Use of cancer chemopreventive phytochemicals as antineoplastic agents. The Lancet Oncology, Vol. 6, No. 11,pp. 899-904.
Dziuba, B., Dziuba, M. (2014) New milk protein-derived peptides with potential antimicrobial activity: An approach based on bioinformatic studies. International Journal of Molecular Sciences, Vol. 15, No. 8,pp. 14531-45.
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S.E., Wilkins, M.R., Appel, R.D., Bairoch, A. (2005) Protein identification and analysis tools on the ExPASy server. Springer.
Guralp, S.A., Murgha, Y.E., Rouillard, J.-M., Gulari, E. (2013) From design to screening: a new antimicrobial peptide discovery pipeline. PLoS One, Vol. 8, No. 3, p. e59305.
Habiba, U., Ahmad, M., Shinwari, S., Sultana, S., Shinwari, Z.K., Zafar, M. (2016) Antibacterial and antifungal potential of himalayan medicinal plants for treating wound infections’, Pakistan Journal of Botany, Vol. 48, No. 1,pp. 371-5.
Hammami, R., Hamida, J.B., Vergoten, G., Fliss, I. (2009) PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Research, Vol. 37, No. suppl 1,pp. D963-D8.
Hammami, R., Zouhir, A., Le Lay, C., Hamida, J.B., Fliss, I. (2010) BACTIBASE second release: a database and tool platform for bacteriocin characterization. Bmc Microbiology, Vol. 10, No. 1,p. 1.
Hancock, R.E., Hilpert, K., Cherkasov, A., Fjell, C. (2015) Small cationic antimicrobial peptides. Google Patents.
Igarashi, Y., Heureux, E., Doctor, K.S., Talwar, P., Gramatikova, S., Gramatikoff, K., Zhang, Y., Blinov, M., Ibragimova, S.S., Boyd, S. (2009) PMAP: databases for analyzing proteolytic events and pathways. Nucleic Acids Research, Vol. 37, No. suppl 1, pp. D611-D8.
Keil, B. (1986) Proteolysis Data Bank: specificity of alpha-chymotrypsin from computation of protein cleavages. Protein Sequences & Data Analysis, Vol. 1, No. 1, pp. 13-20.
Keil, B. (1992) Essential substrate residues for action of endopeptidases. Specificity of Proteolysis, Springer, pp. 43-228.
Krishnan, R.J., Nair, S.R. (2016) Preliminary Study on the Antibacterial Activity of Six Medicinal Plants against Two Naso-Pharyngeal Pathogens—Streptococccus pyogenes and Pseudomonas aeruginosa. American Journal of Plant Sciences, Vol. 7, No. 06,p. 907.
Mor, A., Radzishevsky, I. (2015) Novel antimicrobial agents. Google Patents.
Ortega‐Ramirez, L.A., Rodriguez‐Garcia, I., Leyva, J.M., Cruz‐Valenzuela, M.R., Silva‐Espinoza, B.A., Gonzalez‐Aguilar, G.A., Siddiqui, M.W., Ayala‐Zavala, J.F. (2014) Potential of medicinal plants as antimicrobial and antioxidant agents in food industry: a hypothesis. Journal of Food Science, Vol. 79, No. 2,pp. R129-R37.
Pushpanathan, M., Gunasekaran, P., Rajendhran, J. (2013) Antimicrobial peptides: versatile biological properties. International Journal of Peptides, Vol. 2013.
Sahebrao, K.R., Deepak, L.M. (2014) World Journal of Pharmaceutical Research.
Smith, P.A., Romesberg, F.E. (2007) Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nature Chemical Biology, Vol. 3, No. 9, pp. 549-56.
Taniguchi, M., Ochiai, A., Takahashi, K., Nakamichi, S.i., Nomoto, T., Saitoh, E., Kato, T., Tanaka, T. (2016) Effect of alanine, leucine, and arginine substitution on antimicrobial activity against Candida albicans and action mechanism of a cationic octadecapeptide derived from á‐amylase of rice. Peptide Science.
Tyagi, A., Kapoor, P., Kumar, R., Chaudhary, K., Gautam, A., Raghava, G. (2013) In silico models for designing and discovering novel anticancer peptides. Scientific reports, Vol. 3.
Waghu, F.H., Gopi, L., Barai, R.S., Ramteke, P., Nizami, B., Idicula-Thomas, S. (2014) CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Research, Vol. 42, No. D1,pp. D1154-D8.
Wang, G., Li, X., Wang, Z. (2009) APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Research, Vol. 37, No. suppl 1, pp. D933-D7.
Wang, Z., Wang, G. (2004) APD: the antimicrobial peptide database. Nucleic Acids Research, Vol. 32, No. suppl 1, pp. D590-D2.
Zasloff, M. (2002) Antimicrobial peptides of multicellular organisms. Nature, Vol. 415, No. 6870,pp. 389-95.
Zorofchian Moghadamtousi, S., Abdul Kadir, H., Hassandarvish, P., Tajik, H., Abubakar, S., Zandi, K. (2014) A review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed ResearchInternational, Vol. 2014.


