Laboratory of Microbiology and Experimental Medicine, Department of Zoology,
University of Gour Banga, Malda-732103, West Bengal, India
Corresponding author email: samtropmed@gmail.com
Article Publishing History
Received: 18/11/2023
Accepted After Revision: 24/03/2024
Manifold types of environmental samples have been known to be contaminated with potential bacterial pathogens stretching the global world with several infections in humans. Among six samples including five environmental samples and one food sample were taken up for in vitro study. Applying the disc diffusion method using 10 antibiotics, the isolated bacterial susceptibility to antibiotics was performed and MAR (multiple antibiotic resistance) index was calculated. All the bacterial isolates were sensitive to AK (amikacin), CIP (ciprofloxacin), VA (vancomycin), TE (tetracycline), MRP (meropenem), and IPM (imipenem), and resistant to AMP (ampicillin). The calculated MAR index of the isolates varied from 0.1 to 0.3, where the value was 0.3 for Bacillus sp, Bacillus cereus, and Bacillus subtilis.
In silico molecular docking was also performed to know the binding affinity of two antibiotics IMP and AK against bacterial target proteins (gyrase B and sortase A), wherein the binding energies ranged from -5.7 to -6.8 kcal/mol. The current findings revealed the emergence of antibiotic resistance among bacterial strains in environmental and food samples as well as provided information on bacterial contamination of environmental and food samples. The present study also determined the antibiotic susceptibility pattern and the MAR index to know the source environmental quality. Therefore, routine surveillance of environmental and food samples is warranted to monitor antibiotic resistance of the individual bacterial strains associated.
Cotton Mask, Notes, Bacteria, Antibiotic Resistance, Molecular Docking.
Nandi , Majumdar G, Karmakar R, Mandal J, Sarkar B, Paul A, Mandal A, Das M, Mandal S. Antibiotic Resistance of Bacterial Isolates from Food and Environment: In Vitro and In Silico Analysis. Biosc.Biotech.Res.Comm. 2024;17(1).
Nandi , Majumdar G, Karmakar R, Mandal J, Sarkar B, Paul A, Mandal A, Das M, Mandal S. Antibiotic Resistance of Bacterial Isolates from Food and Environment: In Vitro and In Silico Analysis. Biosc.Biotech.Res.Comm. 2023;17(1). Available from: <a href=”https://shorturl.at/gtSX7“>https://shorturl.at/gtSX7</a>
INTRODUCTION
Microorganisms are found to be present everywhere (including soil, water, air even the human body) and they are microscopic single-celled, prokaryotic organisms (Vidyasagar et al., 2015). It is very important and imperatively necessary to characterize the microbiome of various environments inclusive of indoor surfaces as people spend most of the time in an indoor environment (Andualem et al., 2019). Among all indoor environments, laboratories are relatively safe but may be contaminated with bacteria (Zhu et al., 2020).
Consequent upon the outbreak of the dreaded COVID-19, the face masks are used on a large scale as they have good effects for averting seasonal virus-causing diseases (Enaigbe et al., 2021). It may serve as a substrate for microbial growth by creating the humid habitat released by breathing, coughing, and sneezing. The reuse of cotton masks, poor filtration, and improper sanitization give rise to the possibility of viral and bacterial transmission (Enaigbe et al., 2021).
Computer keyboards and mouse are the high-touch surfaces, which may act as a vehicle for the transmission of potentially pathogenic bacteria. Most computer keyboards are contaminated with pathogenic bacteria including Gram-positive bacteria, such as Staphylococcus sp. and coagulase-negative bacteria such as Enterobacter sp. Different locations (laboratories, institutes, hospitals) represent various kinds of potential pathogenic bacteria (Staphylococcus aureus, Staphylococcus haemolyticus, and Escherichia coli (Nazeri et al., 2019).
Currency notes pose a great threat to the public health as it is the most frequently passed item and also act as a vector for disseminating potentially pathogenic microorganisms. Cross-contamination by regular handling of money with poor sanitation practices cause badly the risk of infection by multi-drug resistant bacterial strains. Staphylococcus sp., E. coli and Pseudomonas sp. are the most commonly found species isolated from different note samples (Abdalrahaman et al., 2020).
Pickling is a traditional method for preserving food by fermentation with the addition of salt, jaggery, oil, and various spices. Multiple ancient civilizations, such as Indians, Chinese, and Egyptians used pickling for food preservation. Pickles are a great source of potential lactobacilli, but poor hygiene and improper storage may be responsible for causing a great risk of high contamination (Behera et al., 2020).
Bacterial antibiotic resistance is nowadays one of the utmost obstacles in health care crisis in (Li et al., 2023). Most of the antibiotics become ineffective against bacteria, since bacteria change themselves by employing various strategies including efflux pump, restrict the access of entryways of antibiotics, destruction of drugs using bacterial enzymes, modification of binding sites of antibiotics, and become resistant (Vila et al., 2020). For clearer close up of antibiotic and bacterial enzymes interaction, in silico studies have been very much effective to understand the mode of binding and interaction for the inhibition of bacterial enzyme responsible for pathogenicity and resistance (Budama-Kilinc et al., 2023; Majumdar and Mandal., 2024; Mandal and Mandal., 2024).
Therefore, from this study different pathogenic bacterial presence were evaluated in our daily usable items as well as food items, and antibiotic resistance profiles were determined. Also, two antibiotics which were used to check the antibiotic resistance profile of bacteria were used for in silico molecular docking study against two bacterial pathogenicity related proteins to analyse the mode of ligand-protein interaction.
MATERIAL AND METHODS
A total six swab samples (five environmental: computer keyboard, currency note, face masks (n=2), and one food: pickle) were collected. Each of the samples was inoculated into the nutrient broth (Hi-Media, India) and MRS broth (Hi-Media, India), and incubated at 37°C for 24 h. From each sample, a loop full of broth culture was taken and then streaked on nutrient agar, MRS agar, and cetrimide agar plate (Hi-Media, India), and incubated for 24 h at 37°C. Nutrient agar stabs and MRS agar stabs were used to preserve single and morphologically discrete bacterial colonies appeared on the plate after subcultures. The isolated bacteria were identified by following gram-staining, gram-reaction, and biochemical tests (TSI, indole, catalase, oxidase, citrate, mannitol salt agar) (Al-Dhabaan., 2019).
Table 1. Biochemical test results for the isolated bacteria from food and environmental samples
| Sl no.
|
Isolated
Bacterial code |
Media | Gram staining | Gram reaction (KOH test) | Biochemical test | ||||||||
| Property | CS | SP | DNS | TSI | CIT | CAT | OXI | MAN | IND | ||||
| 1. | 701 M | NA | positive | rod | + | Non-sticky | + | P/Y | – | – | – | – | – |
| 2. | 702M | NA | positive | rod | – | Non-sticky | – | P/Y | + | + | – | – | – |
| 3. | 703 M | NA | positive | round | – | Non-sticky | – | Y/Y | + | + | – | + | – |
| 4. | 704 B | NA | positive | rod | + | Non-sticky | + | P/Y | – | + | – | – | – |
| 5. | 705 C | NA | positive | rod | + | Non-sticky | + | P/Y | + | + | + | – | – |
| 6. | 706 P | NA | positive | rod | + | Non-sticky | – | Y/Y | + | + | + | – | – |
| 7. | 701 M1 | MRS | positive | round | – | Non-sticky | – | Y/Y | – | + | + | – | – |
| 8. | 702 M1 | MRS | positive | round | – | Non-sticky | – | Y/Y | – | + | + | – | – |
| 9. | 703 M1 | MRS | positive | round | – | Non-sticky | – | Y/Y | + | + | + | – | – |
| 10. | 704 B1 | MRS | positive | round | – | Non-sticky | – | Y/Y | – | + | + | – | – |
| 11. | 705 C1 | MRS | positive | round | – | Non-sticky | – | P/Y | + | + | + | – | – |
| 12. | 701 M2 | CA | positive | rod | + | Non-sticky | + | P/Y | + | + | – | – | – |
| 13. | 702M2 | CA | positive | rod | – | Non-sticky | – | P/Y | – | + | + | – | – |
| 14. | 703 M2 | CA | positive | round | – | Non-sticky | – | P/Y | – | – | – | + | – |
| 15. | 704 B2 | CA | positive | rod | – | Non-sticky | – | P/Y | + | + | + | – | – |
| 16. | 705 C2 | CA | positive | rod | – | Non-sticky | – | P/Y | – | – | – | – | – |
| 17. | 706 P2 | CA | positive | rod | + | Non-sticky | + | Y/Y | – | + | – | – | – |
CAT: Catalase; CIT: citrate; CS: cell shape; DNS: DNase; IND: indole; MAN: mannitol; OXI: oxidase; SI: source of isolates, SP: Spore; TSI: triple sugar iron; NA: nutrient agar; MRS: de Man Rogosa Sharpe agar; CA: cetrimide agar.
The antibiotic susceptibility test was performed for the isolated bacteria, using nutrient agar plate. Pure culture of different bacterial isolates (using the nutrient broth cultures) was swabbed on a different nutrient agar plate. Antibiotic discs: amikacin (AK: 30 μg/disc), ciprofloxacin (CIP: 10 μg/disc), ceftriaxone (CTR), nalidixic (NA), vancomycin (VA: 30 μg/disc), ampicillin (AMP: 10 μg/disc), amoxyclav (AMC), tetracycline (TE: 30 μg/disc), meropenem (MRP: 10 μg/disc) and imipenem (IPM: 100 μg/disc) were placed on each agar plate, which were then incubated at 37°C for 24 h. After incubation, isolates showed zone diameter of inhibition (ZDI) around each antibiotic disc and the results were interpreted following the criteria of the Clinical and Laboratory Standards Institute (CLSI, 2011); the isolated bacteria were categorized as resistant, sensitive, or intermediately susceptible.
Table 2. Identity of isolated bacteria from different environmental and food samples
| Bacterial isolates | Source of isolation | Identity |
| LMEM 701 M | Mask (LMEM-M) | Bacillus cereus |
| LMEM 702 M | Listeria monocytogenes | |
| LMEM 703 M | Staphylococcus aureus | |
| LMEM 701 M1 | Mask (LMEM-M1) | Staphylococcus epidermidis |
| LMEM 702 M1 | Staphylococcus epidermidis | |
| LMEM 703 M1 | Micrococcus luteus | |
| LMEM 701 M2 | Mask (LMEM-M2) | Bacilluscereus |
| LMEM 702 M2 | Corynebacterium sp. | |
| LMEM 703 M2 | Staphylococcus aureus | |
| LMEM 704 B | Computer keyboard | Corynebacterium diphtheriae |
| LMEM 704 B1 | Staphylococcus epidermidis | |
| LMEM 704 B2 | Corynebacterium sp. | |
| LMEM 705 C | Note | Bacillus megaterium. |
| LMEM 705 C1 | Micrococcus luteus | |
| LMEM 706 P | Pickle | Bacillus subtilis |
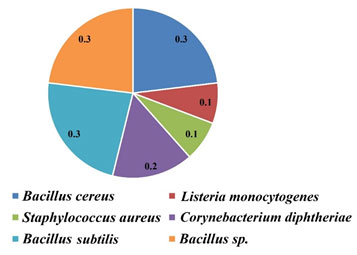
MAR index is the ratio between the number of antibiotics for which the bacterial strain had a resistance and the total number of antibiotics exposed to the particular bacterial strain at the time of the antibiotic susceptibility test (Krumperman., 1983). The MAR index of <0.2 was indicated low risk, and ≥0.2 was indicated as high risk.
Two 3-D structures of bacterial proteins, ATP binding domains of S. aureus gyrase B (PDB ID: 5CPH) and Listeria monocytogenes sortase A (PDB ID: 5HU4), responsible for pathogenicity were retrieved from RCSB protein data bank (https://www.rcsb.org/) and used as the targets. 3-D structures two antibiotics IMP (PubChem CID:104838) and AK (PubChem CID:37768) were retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov/), and used as the ligands. Both the protein and ligand molecules were refined, and optimized with the help of UCSF Chimera software version 1.15 (https://www.cgl.ucsf.edu/chimera/), by removing solvent, ions, and ligand molecules from the protein structure, and adding Gasteiger charges during ligand preparation.
Table 3. MAR indices of isolated bacteria from different sources
| Isolated bacteria | MAR index | Risk factor |
| Bacillus cereus | (3/10) = 0.3 | Highly risk |
| Listeria monocytogenes | (1/10) = 0.1 | — |
| Staphylococcus aureus | (1/10) = 0.1 | — |
| Corynebacterium diphtheriae | (2/10) = 0.2 | Moderately risk |
| Bacillus sp. | (3/10) = 0.3 | Highly risk |
| Bacillus subtilis | (3/10) = 0.3 | Highly risk |
Two antibiotics were docked with ATP binding domains of S. aureus gyrase B (5CPH) and L. monocytogenes sortase A (5HU4), using UCSF Chimera inbuilt AutoDock Vina (https://www.cgl.ucsf.edu/chimera/). The active binding sites of the targets were defined using grid box centre: X=15, Y=11, Z= -2, size: X=52, Y=52, Z=52, for protein ATP binding domains of S. aureus gyrase B, and grid box centre: X=12, Y=18, Z= 18, size: X=52, Y=52, Z=52, for L. monocytogenes sortase A. Based upon the previous studies, binding energies of molecular docking ≤ -6.5 kcal/mol were considered for favourable binding and protein inhibition by the ligands (Majumdar and Mandal., 2023; Mandal and Mandal., 2021).
Table 4. Molecular docking results of two bacterial proteins Staphylococcus aureus ATP binding domain of GyrB (5CPH) and Listeria monocytogenes sortase A (5HU4) with two antibiotics imipenem and amikacin
| Protein and ligand | Involved amino acid in hydrogen bond formation with Distance
(Å) |
Involved amino acid in Van der Waals interaction | Involved amino acid in hydrophobic interaction | Binding energy (kcal/mol) |
| 5CPH and imipenem | Glu58 (2.26) | Ile51, Ser55, Thr173, Pro87, Asn54, Asp81, Ser55, Asp57, Gly85, Gly83 | Ile86, Ile102, Ile175 | -6.2 |
| 5CPH and amikacin | Ser55 (2.89), Gly85 (3.05), Asn54 (2.14), Ile102 (2.67), Ser128 (2.29), Asp57 (2.41), Glu58 (2.20) | Ser129, Leu103, Asp81, Thr173, Ile86, Gly83, Pro87, Leu60, Ala61, Tyr63, Arg84 | -6.8 | |
| 5HU4 and imipenem | Arg126 (2.33), Glu98 (2.44), Glu100 (2.35), Asp97 (2.55) | Thr28, Thr38, His56, Gly55, Met40, Ile104, Thr99, Val101, Ile96, Val103, Ile115 | Ala54, Leu33 | -5.7 |
| 5HU4 and amikacin | Asn21(3.15), Asp19 (3.71), Phe148(3.49), Ala35 (3.75) | Phe148,Pro149, Ser150, Ser150, Ser12, Leu23, Leu22, Ala35, Val20, Pro67, Leu64, Ser62, Asp19 | -5.9 |
RESULTS
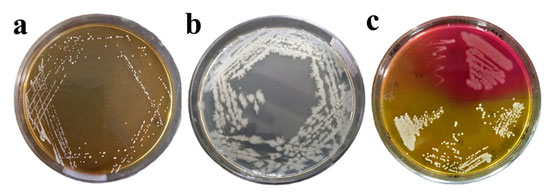
Among the isolated bacteria, 7 were round-shaped and the other 10 were rod-shaped (Table 1). In the DNase test, the strains (701M, 704B, 705C, 701M2 and 706P2) showed a transparent zone. In Triple Sugar Iron (TSI) test,10 strains (701M, 702M, 704B, 705C, 705C1, 701M2, 702M2, 703M2, 704B2, 705C2) showed acid butt (yellow) and alkali slant (pink); no strains were found positive for CO2 and H2S production. Two bacterial strains (703M and 703M2) showed a positive result for the mannitol test. No strains were found positive for the gram reaction test. The biochemical test results for the isolated bacteria is shown in Table 1. Based upon the cultural characteristics (colony morphology, pigment production), gram staining (cell shape), and biochemical test including TSI test results, and DNase test patterns of the environmental and food bacteria, their identities are represented in Table 2 and Fig. 1.
Figure 1: (a) Micrococcus luteus in MRS agar plate. Isolated characteristic Circular, slightly yellow, convex, and smooth, (b) Bacillus cereus in nutrient agar plate. Isolated characteristic grey-white with a less wavy wedge, (c) Mannitol salt agar plate, isolated bacteria from the NA and CA agar plate cultured to determine the particular cocci strain.
All the bacterial strains cultured in the nutrient plate were sensitive to AK, CIP, VA, TE, MRP, and IPM and resistant to AMP. However bacterial strain 701M, 706P, 704B, and 705C were resistant to CTR, in addition, while 702M and 703M were sensitive and intermediately sensitive, respectively. Although strain 701M, 706 and 705C showed resistance to AMC, bacterial isolates 702M and 703M were sensitive, and the isolate 704B was intermediately sensitive. The comparison of all the antibiotic susceptibility or resistances (Fig. 2 and Fig 3).
Figure 2: (a) Antibiotic susceptibility test results for mask samples and (b) Antibiotic susceptibility
test results for pickle, notes and keyboard. Abbreviations of antibiotics are mentioned in the text.
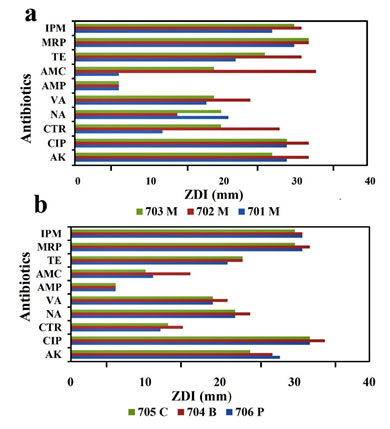
Figure 3: Antibiotic susceptibility test results following the by disc
diffusion methods of mask samples.

In the case of MRS agar, all the strains were highly susceptible to antibiotics and the Zone Diameter of Inhibition (ZDI) was >40 mm. The MAR indices of isolated bacteria are shown in Table 3 and Fig. 4.
Figure 4: MAR indices of isolated bacteria
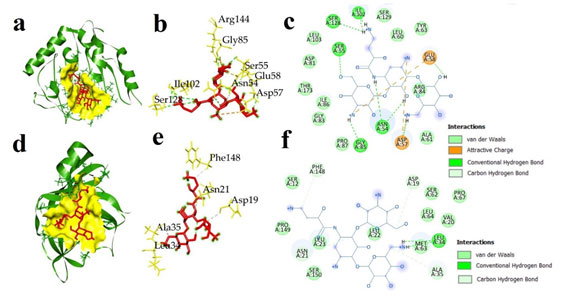
The interaction between IMP and AK against two pathogenic bacterial proteins: ATP binding domains of S. aureus gyrase B (5CPH) and L. monocytogenes sortase A (5HU4) were considered to know the mechanism of binding of antibiotics with target proteins. AK showed lowest binding energy of -6.8 kcal/mol against S. aureus gyrase B ATP binding domain, and -5.9 kcal/mol against L. monocytogenes sortase A. The IMP had binding energies of -6.2 kcal/mol against S. aureus gyrase B ATP binding domain and -5.9 against L. monocytogenes sortase A (Table 4). During docking, several types of interactions were formed between protein and antibiotics that help in the stability of the complexes and determine binding affinity.
IMP interacted with S. aureus gyrase B ATP binding domain and formed one hydrogen bond and salt bridge with Glu58 (2.26 Å), ten Van der Waals interactions with Ile51, Ser55, Thr173, Pro87, Asn54, Asp81, Ser55, Asp57, Gly85, and Gly83, and three hydrophobic interactions in the form of π-alkayl with Ile86, Ile102, and Ile175 (Fig. 5). AK interacted with S. aureus gyrase B, displaying two electrostatic interactions with Glu58, Asp57, seven conventional hydrogen bonds with Ser55 (2.89 Å), Gly85 (3.05 Å), Asn54, (2.14 Å), Ile102 (2.67 Å), Ser128 (2.29 Å), Asp57 (2.41 Å), and Glu58 (2.20 Å), one carbon hydrogen bond with Asn54, and a total eleven Van der Waals interactions with Ser129, Leu103, Asp81, Thr173, Ile86, Gly83, Pro87, Leu60, Ala61, Tyr63, and Arg84 (Fig. 6).
Figure 5: Docked complex of imipenem and two bacterial proteins S. aureus ATP binding domain of GyrB (5CPH) and L. monocytogenes sortase A (5HU4) (a) 3D docked complex of (5CPH) with imipenem (b) 3D interaction of 5CPH with imipenem (c) 2D representation of interacted amino acids of 5CPH with imipenem (d) 3D docked complex of 5HU4 with imipenem (e) 3D interaction of 5HU4 with imipenem (f) 2D representation of interacted amino acids of 5HU4 with imipenem.
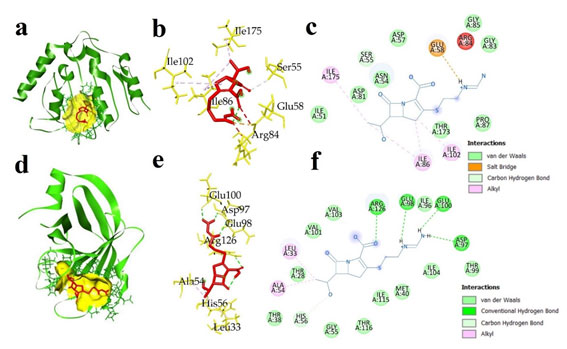
L. monocytogenes sortase A with IMP showed four hydrogen bonds with Arg126 (2.33 Å), Glu98 (2.44 Å), Glu100 (2.35 Å), Asp97 (2.55 Å), ten Van der Waals interactions with Phe148, Pro149, Ser150, Ser12, Leu23, Leu22, Ala35, Val20, Pro67, Leu64, Ser 62, Asp19, and two π-alkayl interactions with Ala54 and Leu33 (Fig. 5). AK formed four hydrogen bonds (carbon hydrogen) with Asn21 (3.15 Å), Asp19 (3.71 Å), Phe148 (3.49 Å), Ala35 (3.75 Å), one conventional hydrogen bond with Leu34 (2.20 Å) and a total of 13 Van der Waals interactions with amino acids Phe148, Pro149, Ser150, Ser150, Ser12, Leu23, Leu22, Ala35, Val20, Pro67, Leu64, Ser62, and Asp19 (Fig. 6).
Figure 6: Docked complex of amikacin and two bacterial proteins S. aureus ATP binding domain of GyrB (5CPH) and L. monocytogenes sortase A (a) 3D docked complex of (5CPH) with amikacin (b) 3D interaction of 5CPH with amikacin (c) 2D representation of interacted amino acids of 5CPH with amikacin (d) 3D docked complex of 5HU4 with amikacin (e) 3D interaction of 5HU4 with amikacin (f) 2D representation of interacted amino acids of 5HU4 with amikacin.
DISCUSSION
Contamination with bacteria in food and environment is common and isolation of bacterial strains from food such as homemade pickle and environment samples are not unknown. Different authors from different parts of the world identified various types of bacteria from the environment and food. According to Gund et al. (2021), some species such as Staphylococcus epidermidis, Staphylococci sp., M. luteus, Rothia dentocariosa, Streptococcus oralis, and Bacillus sp. were found in a surgical mask. Nightingale et al. (2022) stated that face masks can contain E. coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and S. aureus. But in the current study, bacterial strains in cotton masks such as Bacillus cereus, S. aureus, S. epidermidis, L. monocytogenes, Micrococcus luteus and Corynebacterium sp. were found. All the bacterial isolates showed sensitivity against 9 antibiotics and resistance to one.
In accordance with the report of Al-Akeedi et al., 2021, Bacillus sp., Corynebacterium sp., micrococcus sp., Staphylococcus sp., B. cereus, Proteus sp., P. aeruginosa, Streptococcus viridians, and E. coli were isolated from different computer keyboards. As per the current study, Corynebacterium diphtheria and Staphylococcus epidermidis were the major bacteria isolated from computer keyboards. In pursuance of the report of Abba and Okoye (2022), bacterial strains such as S. aureus, Escherichia coli, Pseudomonas sp., S. epidermidis, Klebsiella sp. and Bacillus sp. were identified in Naira notes.
But our current study showed the existence of Bacillus sp. and M. luteus in Indian currency, and these two strains showed sensitivity against 9 antibiotics and resistance against to one. Uchino et al. (2020) reported the presence of Salmonella sp., S. aureus, B. cereus and E. coli food spoilage strains that mainly contaminate pickle. In the current study, bacteria including Bacillus subtilis and Bacillus megaterium were isolated from the vegetable pickle. Another study, by Aljahani (2020), revealed the isolation of E. coli, Salmonella enterica and L. monocytogenes in various pickle samples.
cereus is widespread and commonly found in soil, water, and food. It causes foodborne illnesses including vomiting and diarrhoea. It is generally resistant to β-lactam antibiotics such as ampicillin, penicillin, and amoxicillin, but susceptible to vancomycin and erythromycin (Uchino et al., 2020). This bacterial strain was found in eye cosmetics (Nandi and Mandal, 2016) and also found in the face mask, which indicated high contamination. S. aureus is a Gram-positive potential pathogenic bacterium that causes skin and tissue infections in humans. By genetic mutations, it becomes multi-drug resistant. The toxic elements of S. aureus cause gastrointestinal symptoms (Uchino et al., 2020).
B. subtilis also known as hay bacillus is a gram-positive bacterium mainly found in soil, the gastrointestinal tract of ruminants, humans, and marine sponges. This bacterium is sensitive to TET and VM but resistant to streptomycin (Colom et al., 2021). B. megaterium is an aerobic spore-forming Gram-positive bacteria widespread in nature (John, 2020). In our study, this bacterial strain was found in currency notes. C. diphtheriae, the causative agent of diphtheria, which is the main reason for thousands of deaths per year. This bacterial strain is a toxigenic bacterium, but if we review our study this bacterial strain was found in keyboard (Ott et al., 2022). Foodborne pathogen L. monocytogenes a causative agent for listeriosis.
This gram-positive bacterium was first isolated from rabbit liver (Disson et al., 2021). Though it is a foodborne bacterium, we found this strain in the mask sample, which was also found in eye cosmetics (Nandi and Mandal., 2016). Gram-positive bacteria, according to Li et al., (2021), M. luteus the main pathogen for nosocomial infections and in the present study, we found this strain in mask and currency notes. Another bacterium that can make biofilm in the skin, especially the nasal area is S. epidermidis, which was found in mask and keyboard samples (Ortega-Peña et al., 2022).
From the molecular docking studies, ATP binding domain of S. aureus GyrB with IMP showed -6.2 kcal/mol and with AK -6.8 kcal/mol of binding energies, and the L. monocytogenes sortase A with IMP and AK showed binding energies of -5.7 and -5.9 kcal/mol. ATP binding domain of S. aureus GyrB docked complex with AK, showed intense lower binding energy as compared to others (Terefe and Ghosh., 2022). In all the four complex as formed by different interactions mainly hydrogen bonds, Van der Waals interactions and hydrophobic interactions, but the inhibition potency was highest in case of ATP binding domain of S. aureus GyrB and AK according to docking results (Santha et al., 2022).
Less interaction energy might be associated with the formation of hydrogen bonds (Li et al., 2023). The docking analysis provided the insights of the mechanism of binding of antibiotic to their bacterial counterpart responsible for infection, and the protein-ligand interaction suggested the amikacin as well as imipenem both of them were effective inhibitors, as also supported by the in vitro study.
CONCLUSION
All the bacterial isolates were sensitive to AK, CIP, VA, TE, MRP and IPM and resistant to AMP. Isolated bacteria such as C. diphtheria, S. epidermidis, and L. monocytogenes notable in the current study. The keyboard can cause the spreading of contagious diseases, so the keyboard must be disinfected regularly. Cash notes are the most variable things in the world. So, every time someone used those notes, must sanitize their hands properly. Vegetable pickle is a preserved food, which was found to be contaminated with bacteria such as B. subtilis that might potentially cause infection. So, pickle should be prepared carefully following hygienic ways, and should be stored in properly sterilized containers.
Bacteria such as Bacillus cereus, C. diphtheriae, and B. subtilis which were isolated from the tested samples had MAR index ≥0.2, which indicated the bacterial origin from highly antibiotic polluted sources. Besides that, isolated L. monocytogenes and S. aureus showed comparatively lower MAR index, which was 0.1 indicating their origin from an environment with moderately antibiotic contamination. In silico studies further revealed the effectiveness of the antibiotics (AK and IMP) against bacterial target proteins, which implied possible cause of bacterial susceptibility.
Conflict of interest : The authors have no conflict of interest.
Funding: Authors did not receive any funding for this work.
Data Availability: Data will be available on request
REFERENCES
Al-Akeedi, J.M., Hassan, A.H. and Alhiti, M.A.J., 2021. Bacterial contamination of computer keyboards in pharmacy college / Baghdad University and Al-Rasafa internet centers. Annals of the Romanian Society for Cell Biology, 25(6), pp.9994-9998.
Al-Dhabaan, F.A., 2019. Morphological, biochemical and molecular identification of petroleum hydrocarbons biodegradation bacteria isolated from oil polluted soil in Dhahran, Saud Arabia, Saudi Journal of Biological Sciences, 26(6), pp.1247-1252.
Ali, A.H., Abdalrahman, A.T., Ahmed, A.M., Hajhamed, D.H.A., Abdalrazig, M.B.A., Mohammed M.H.F., Hamad, M.N.M. and Bahar, M., 2020. Isolation of the pathogenic bacteria from banknotes and coins in Khartoum City Pre-COVID-19 era, Sudan. Saudi Journal of Biomedical Research, 5(12), pp.363-367.
Aljahani, A.H., 2020. Microbiological and physicochemical quality of vegetable pickles. Journal of the Saudi Society of Agricultural Sciences, 19(6), pp.415-421.
Andualem, Z., Gizaw, Z., Bogale, L. and Dagne, H., 2019. Indoor bacterial load and its correlation to physical indoor air quality parameters in public primary schools. Multidisciplinary Respiratory Medicine, 14(2), pp.1-7.
Behera, S.S., Sheikha, A.F.E., Hammami R. and Kumar, A., 2020. Traditionally fermented pickles: how the microbial diversity associated with their nutritional and health benefits? Journal of Functional Foods, 70, 103971.
Budama-Kilinc, Y., Gok, B., Cetin Aluc, C. and Kecel-Gunduz, S., 2023. In vitro and in silico evaluation of the design of nano-phyto-drug candidate for oral use against Staphylococcus aureus. PeerJ, 11(e15523), pp.1-27.
Clinical and Laboratory Standards Institute (CLSI) (2011). Performance standards for antimicrobial susceptibility testing, 21st informational supplement M100S21. CLSI, Wayne, Pa
Colom, J., Freitas, D., Simon, A., Brodkorb, A., Buckley, M., Deaton, J. and Winger A.M., 2021. Presence and germination of the probiotic Bacillus subtilis DE111 in the human small intestinal tract: A randomized, crossover, double-blind, and placebo-controlled study. Frontiers in Microbiology, 12, 715863.
Devi, J., Sood, S., Vidyasagar, V. and Singh, Y., 2015. Inheritance of bacterial wilt resistance and performance of horticultural traits in bell pepper (Capsicum annuum var. grossum). Indian Journal of Agricultural Sciences, 85(11), pp.1498-1503.
Disson, O., Moura, A. and Lecuit, M., 2021. Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends in microbiology, 29(9), pp.811-822.
Gund, M., Isack, J., Hannig, M., Thieme-Ruffing, S., Gärtner, B., Boros, G. and Rupf, S., 2021. Contamination of surgical mask during aerosol-producing dental treatments. Clinical Oral Investigations, 25(5), pp.3173–3180.
Irodi C.C., Enaigbe, A.A. and Akpoka O. A., 2021. Bacterial contaminations of used face mask collected from different clinical sections in a university teaching hospital during COVID-19 pandemic crises in Nigeria. Bacterial Empire, 4(1), pp.8-10
Krumperman, P.H., 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and Environmental Microbiology, 46(1), pp.165–170.
Li, H., Komori, A., Li, M., Chen, X., Yang, A. W. H., Sun, X., Liu, Y., Hung, A., Zhao, X. and Zhou, L., 2023. Multi-ligand molecular docking, simulation, free energy calculations and wavelet analysis of the synergistic effects between natural compounds baicalein and cubebin for the inhibition of the main protease of SARS-CoV-2. Journal of Molecular Liquids, 374 (121253), pp.1-19.
Li, T., Wang, Z., Guo, J., de la Fuente-Nunez, C., Wang, J., Han, B., Tao, H., Liu, J. and Wang, X. 2023. Bacterial resistance to antibacterial agents: Mechanisms, control strategies, and implications for global health. The Science of The Total Environment, 860, p.160461.
Li, Y., Sun, Z.Z., Rong, J.C. and Xie, B.B., 2021. Comparative genomics reveals broad genetic diversity, extensive recombination and nascent ecological adaptation in Micrococcus luteus. BMC genomics, 22(1), p.124.
Majumdar, G. and Mandal, S., 2023. Exploring the Inhibitory role of Persicaria hydropiper bioactive compounds against 2KID protein associated with Staphylococcus aureus biofilm formation: Molecular docking and pharmacological property analysis. Research Journal of Pharmacy and Technology, 16(7), pp.3189-3194.
Majumdar, G., and Mandal, S., 2024. Evaluation of broad-spectrum antibacterial efficacy of quercetin by molecular docking, molecular dynamics simulation and in vitro studies. Chemical Physics Impact, 8(100501), pp.1-15.
Mandal, M. and Mandal, S., 2021. Molecular docking and dynamics simulation of L-hyoscyamine, eupatorium and alkaloid L27 as potential inhibitors against 3CLpro of SARS-CoV-2. Drug Discovery, 15(36), pp.231-251.
Mandal, M. and Mandal, S., 2024. Discovery of multitarget-directed small molecule inhibitors from Andrographis paniculata for Nipah virus disease therapy: molecular docking, molecular dynamics simulation and ADME-Tox profiling. Chemical Physics Impact, 8 (100493), pp.1-16.
Muramatsu, S., Uchino, M., Sorm, S., Oka, D., Muramatsu, S., Nakajima, T., Sekido M., Nakamura, T., Chay, C. and Mihara, M., 2020. Evaluation of bacterial contamination levels in pickles sold at wet market in Cambodia-Part 2-detection of several food-poisoning bacteria of 48 samples from Phnom Penh. International Journal of Environmental and Rural Development, 11(1), pp.121-126.
Nandi, S. and Mandal, S., 2016. Bacteriological profiling of commercially available eye cosmetics and their antibiotic susceptibility pattern. Translation Biomedicine, 7(3), pp.1-8.
Nazeri, M., Arani, J.S., Ziloochi, N., Delkhah, H., Arani, M.H., Asgari, E. and Hosseini, M., 2019. Microbial contamination of keyboards and electronic equipment of ICU (Intensive Care Units) in Kashan University of medical sciences and health service hospitals. MethodsX, 6, pp.666–671.
Ortega-Peña, S., Rodríguez-Martínez, S., Cancino-Diaz, M.E. and Cancino-Diaz, J.C., 2022. Staphylococcus epidermidis controls opportunistic pathogens in the nose, could It Help to regulate SARS-CoV-2 (COVID-19) infection? life. 12(3), p.341.
Ott, L., Möller, J. and Burkovski, A., 2022. Interactions between the Re-emerging pathogen Corynebacterium diphtheriae and host Cells. International Journal of Molecular Sciences, 23(6), p.3298.
Santha, SSR. and Vishwanathan, AS., 2022. Mechanistic insights into 5-lipoxygenase inhibition by pyocyanin: a molecular docking and molecular dynamics study. Journal of Biomolecular Structure and Dynamics, 40(20), pp.9752–9760.
Terefe, EM. and Ghosh, A., 2022. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of phytochemicals isolated from croton Dichogamus against the HIV-1 reverse transcriptase. Bioinformatics and Biology Insights, 16, (11779322221125605) pp.1-20.
Vila, J., Moreno-Morales, J. and Ballesté-Delpierre, C., 2020. Current landscape in the discovery of novel antibacterial agents. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 26(5), pp.596–603.
Zhu, X., Li, X., Wang, W. and Ning K., 2020. Bacterial contamination screening and interpretation for biological laboratory environments. Medicine in Microecology, 5(1), p.100021.


