1Department of Biological Science, Faculty of Science, King Abdulaziz University, Jeddah, Kingdom of Saudi Arabia
Corresponding author email: ahmadfirozbin@gmail.com
Article Publishing History
Received: 12/10/2020
Accepted After Revision: 14/12/2020
Parkinson’s disease (PD) is second most common motoric neurodegenerative disorder, which affects population above age 65It is characterized by the loss of dopaminergic neurons from substantia and the presence of intracellular. SNCA is a protein encoded by 5 exons with total transcript length of 3041 bps maps on 4q21.3-q22. SNCA gene is considered to be involved in regulation of dopamine release and transport, induces fibrillization of microtubule associated protein tau, and exert neuroprotective phenotype in non-dopaminergic neurons by inhibiting both p53 expression and transactivation of proapoptotic genes leading to decreased caspase-3 activation. Polymorphisms in Alpha-synuclein (SNCA) gene have been associated with Parkinson disease.
In this study, computational analysis of pathogenic SNPs of SNCA gene has been performed to identify and analyze the deletrious SNPs using bioinformatics approach. We obtained pathogenic SNPs data from dbSNP database. We employed consensus tools SIFT, PROVEAN, Condel, PolyPhen-2 to predict deleterious pathogenic nonsynonymous SNPs, Pathogenic mutants A30P (rs104893878) and G51D (rs431905511) shows deleterious by all four tools and three tools respectively. These predicted pathogenic deleterious nsSNPs are expected to have impending functional influence and may contribute in understanding the functional roles of SNCA gene associated with Parkinson disease.
nsSNP, Alpha-synuclein, SNCA, In Silico Analysis.
AlGhanmi E. A. H, Ali H. M, Al-Ghamdi M. K, Firoz A. Analysis of Pathogenic nsSNPs in Human SNCA Gene Associated with Parkinson Disease. Biosc.Biotech.Res.Comm. 2020;13(4).
AlGhanmi E. A. H, Ali H. M, Al-Ghamdi M. K, Firoz A. Analysis of Pathogenic nsSNPs in Human SNCA Gene Associated with Parkinson Disease. Biosc.Biotech.Res.Comm. 2020;13(4). Available from: <a href=”https://bit.ly/39F2zTw”>https://bit.ly/39F2zTw</a>
Copyright © AlGhanmi et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Parkinson’s disease (PD) is second most common motoric neurodegenerative disorder (Mhyre TR et al, 2012; Bisaglia, et al, 2010) which affects 1–2% of the population above age 65 and 4–5% above age 85 (Bisaglia et al,2009).It is characterized by the loss of dopaminergic neurons from substantia and the presence of intracellular (Vilar et al,2008). SNCA is a 14.5 kDa, 140 a.a protein encoded by 5 exons with total transcript length of 3041 bps maps on 4q21.3-q22. The other members of synuclein family are SNCB and SNCG mapped to human chromosome 5q35 and 10q23.2-q23.3 respectively (George,2002). Architecture of SNCA protein reveals the presence of N-terminal region composed of incomplete KXKEGV motifs, extremely hydrophobic NAC domain and highly acidic C-terminal domain8. At physiological conditions, SNCA is believed to be intrinsically disordered monomer or helically folded tetramer10. Oligomeric structure of SNCA is considered as a toxic form but recent observation abolished this hypothesis, (Dettmer et al,2015). Hypotheses exist about toxic structural form of SNCA, but none of them are completely consensual. However, neurotoxic form of SNCA aggregates within neuron and spreads across the anatomically interconnected regions of PD brain through interneuronal transmission using various mechanisms (Recasens et al,2014).
Although SNCA is expressed predominately in brain, it is also expressed in heart, skeletal muscle and pancreas (Lücking& Brice,2000).Molecular function of SNCA is quite ambiguous. Based on its structure, physical properties and several hypotheses for the normal function of SNCA have been proposed. It is considered to be involved in regulation of dopamine release and transport, induces fibrillization of microtubule associated protein tau, and exert neuroprotective phenotype in non-dopaminergic neurons by inhibiting both p53 expression and transactivation of proapoptotic genes leading to decreased caspase-3 activation (Huang, 2018).
However, missense mutations (especially A53T) in (Tang et al,2008).SNCA abolished the neuroprotective effect of SNCA and promote apoptosis by reversing the expression of p53 (da Costa et al,2000). Due to the significant role of SNCA in FPD and other neurodegenerative disorders, premeditated to decipher the molecular evolution of SNCA, which infers the phylogenetic history of synuclein family with the help of its putative orthologs and paralogs. Analysis revealed the sarcopterygian specific origin of SNCA which suggested its lineage specific functional role. On account of this interest, a comparative sequence and structural analysis was performed to estimate the selection and functional constraints on SNCA. Evolutionary rate difference was coupled with structural information to infer potential functional changes and the impact of lineage specific substitutions. In addition, variations in domain topologies were explored by comparative analysis of known functional domains of SNCA protein. In light of the findings, it was hypothesized that the region from 32 to 58 of N-terminal lipid binding domain is the most “critical region” of SNCA from evolutionary, functional and disease pathogenesis perspective. Multiplications of the SNCA gene locus, including duplications and triplications, have been shown to cause autosomal dominant PD in which gene dosage determines severity and latency (Ferese, 2015).
This suggests that 1) the pathologic properties of α-synuclein are not dependent on mutations that alter the α-synuclein protein product, 2) overexpression of wild-type α-synuclein is sufficient to cause disease in a dose-dependent manner and 3) overabundance of α-synuclein may be a common feature of PD wherein smaller increases in α-synuclein, due to increased expression or decreased clearance, may contribute to sporadic disease (Singleton & McCormack,2007). So Alpha-synuclein (SNCA) is considered as the major causative gene involved in the early onset of familial Parkinson’s disease (FPD) characterized by missense mutations reported A30P (;E46K, H50Q, G51D and A53T. SNCA is also deemed to be involved in various other neurodegenerative disorders i.e. Alzheimer’s disease (AD), Lewy bodies’ disease (LBD) and Muscular System Atrophy (MSA) (Kruger,1998, Hashimoto & Masliah,1999 Siddiqui, 2016 ).
MATERIAL AND METHODS
Datasets: The SNPs of the Alpha-synuclein (SNCA) gene were retrieved from the dbSNP database (Sherry ST, 2001). Keyword “Human SNCA” used as our search term. It is filtered by selecting variation class as SNV, function class as missense, clinical significance as pathogenic. Protein sequence of SNCA (uniport id: P37840) was retrieved from uniport database.
Prediction of deleterious and damaging nsSNPs: In order to predict the damaging or deleterious nsSNPs, multiple consensus tools were employed by using online tool VEP (http://www.ensembl.org/Tools/VEP). VEP advantages include, it can predict thousands of SNPs from multiple tools including SIFT, PROVEAN, Condel, and PolyPhen-2, at a time. Pathogenic nsSNP rs-ids were uploaded to VEP tool to get the prediction results
SIFT: The algorithm predicted that the tolerant and intolerant coding base substitution based upon properties of amino acids and homology of sequence (Choi Y, 2015). The tool considered that vital positions in the protein sequence have been conserved throughout evolution and therefore substitutions at conserved alignment position is expected to be less tolerated and affect protein function than those at diverse positions., SIFT predicted substituted amino acid as damaging at default threshold score <0.05, while score ³ 0.05 is predicted as tolerated.
PolyPhen-2: This tool is predicting the structural and functional consequences of a particular amino acid substitution in human protein (Adzhubei, 2010). Prediction of PolyPhen-2 is based on a number of features including information of structural and sequence comparison. The PolyPhen-2 score varies between 0.0 (benign) to 10.0 (damaging). The PolyPhen-2 prediction output categorizes the SNPs into three basic categories, benign (score < 0.2), possibly damaging, (score between 0.2 and0.96), or probably damaging (score >0.96).
Provean: This tool (http://provean.jcvi.org/) uses an alignment-based scoring method for predicting the functional consequences of single and multiple amino acid substitutions, and in-frame deletions and insertions (Choi Y, 2015). The tool has a default threshold score, i.e. -2.5, below which a protein variant is predicted as deleterious, and above that threshold, a protein variant is neutral.
Condel (CONsensus DELeteriousness): This tool evaluates the probability of missense single nucleotide variants (SNVs) deleterious. it computes a weighted average of the scores of SIFT, PolyPhen2, MutationAssessor and FatHMM (Hecht M, 2015).
RESULTS AND DISCUSSION
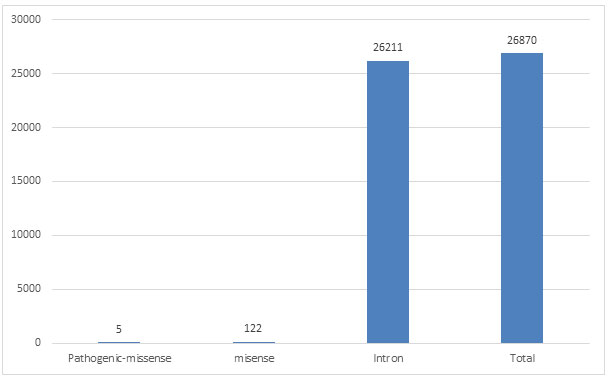
Five SNP-ids of pathogenic nsSNPs mapped in human SNCA gene was downloaded from dbSNP database of NCBI (Table 1), after filtering variation class SNV, function class missense and clinical significance as pathogenic, there were 122 SNP mapped to missense, 26211 SNPs mapped to intron, while 26870 mapped to total SNPs of different variation class (Figure 1). Some rsIDs are associated with multiple SNPs and therefore fall in different classes.
Figure 1: Number of SNPs in different function class of SNCA gene of human from dbSNP database
Predicting deleterious and damaging pathogenic nsSNPs: In order to predict the damaging or deleterious pathogenic nsSNPs multiple consensus tools were employed. Initially, online tool VEP was used (McLaren W, 2016). VEP advantages include: it uses latest human genome assembly GRCh38.p10, and can predict thousands of SNPs from multiple tools including SIFT, Condel, and PolyPhen-2, at a time. 5 nsSNP accession numbers were uploaded to VEP tool and the prediction results were taken on default scores of consensus tools based on sequence and structure homology methods: (a) SIFT (score <-0.5) (b) Polyphen (score >0.96) (c) PROVEAN (score< 2.5) and Condel (score >0.522). In order to get a very high confident nsSNPs impacting structure and function of SNCA gene, 2 nsSNPs (Table 1) are found to be deleterious or damaging by three prediction tools and 1 nsSNP were found to be deleterious by all four tools. These two nsSNPs rs104893878 of mutation A30P and rs431905511 of mutation G51D were further analysed by Netsurf tools for Protein Surface Accessibility and Secondary Structure.
Table 1. Prediction of five pathogenic missense SNPs of SNCA gene using prediction tools such as SIFT, Condel, Polyphen and PROVEAN.
| SNP-ids | AA-Change | SIFT
(score) |
PolyPhen
(score) |
PROVEAN
(score) |
Condel
(score) |
| rs104893878 | A30P | Deleterious
(0) |
Probably damaging (0.996) | Deleterious (-4.536) | Deleterious
(0.906) |
| rs104893875 | E46K | Deleterious
(0.03) |
Benign
(0.003) |
Deleterious (-3.477) | Neutral
(0.364) |
| rs201106962 | H50Q | Tolerated
(1) |
Benign
(0.014) |
Neutral (0.895) | Neutral
(0.001) |
| rs431905511 | G51D | Deleterious
(0.03) |
probably damaging (0.999) | Neutral
(-2.302) |
Deleterious
(0.855) |
| rs104893877 | A53T | Tolerated
(1) |
Benign
(0.01) |
Neutral (1.828) | Neutral
(0.001) |
Studies show a strong evidence about variants are found in SNCA gene involving with Parkinson disease (Jackson, 2003, Stijnen, 2016, Stijnen, 2016). This gene involved in the early onset of familial Parkinson’s disease (FPD) characterized by missense mutations reported A30P, E46K, H50Q, G51D and A53T ((Appel, 2013, Kiely, 2015). Our investigation shows two pathogenic mutants very high confident nsSNPs impacting structure and function of SNCA gene, nsSNPs rs104893878 of mutation A30P found to be deleterious or damaging by three prediction tools and nsSNP rs431905511 of mutation G51D found to be deleterious by all four tools. These two nsSNPs may offer valuable information in selecting SNPs that are expected to have impending functional influence and pathogenicity.
CONCLUSION
Our investigation shows two pathogenic mutants very high confident nsSNPs impacting structure and function of SNCA gene, nsSNPs rs104893878 of mutation A30P found to be deleterious or damaging by three prediction tools and nsSNP rs431905511 of mutation G51D found to be deleterious by all four tools. These two nsSNPs may offer valuable information in selecting SNPs that are expected to have impending functional influence and pathogenicity also eventually may contribute in understanding the functional roles of SNCA gene associated with Parkinson disease.
ACKNOWLEDGMENTS
This work was not supported by any funding agency. We acknowledge with thanks Bioinformatics and Computational Biology Unit at Dept of Biological Sciences, King Abdulaziz University, Jeddah, KSA for providing their support and facilities.
REFERENCES
Adzhubei IA, Schmidt S, Peshkin L,et al (2010) A method and server for predicting damaging missense mutations. Nat Methods. 7(4):248-9.
Appel-Cresswell S, Vilarino-Guell C, Encarnacion M et al (2013). Alpha-synuclein p.H50Q, a novel pathogenic mutation for Parkinson’s disease. 28(6):811-3.
Bisaglia M, Mammi S, Bubacco L (2009). Structural insights on physiological functions and pathological effects of alpha-synuclein. FASEB J.23(2):329-40
Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. (2009). Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 30(8):1237-44.
Choi Y, Chan AP (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 15;31(16):2745-7
Ferese R., Modugno N, Campopiano R, Santilli M. et al, (2015). Four Copies of SNCA Responsible for Autosomal Dominant Parkinson’s Disease in Two Italian Siblings. Parkinson’s disease, 2015, 546462.
Hashimoto, M. & Masliah, E.(1999). Alpha synuclein in Lewy Body Disease and Alzheimer’s Disease. Brain pathology 9, 707–720.
Hecht M., Bromberg & Rost, B. (2015). Better prediction of functional effects for sequence variants. BMC Genomics 16, S1.
HuangYN, Yang LY, Greig NH, Wang YC, Lai CC and Wang JY. (2018). Neuroprotective effects of pifithrin-α against traumatic brain injury in the striatum through suppression of neuroinflammation, oxidative stress, autophagy, and apoptosis. Scientific reports, 8(1), 2368.
Kiely AP, Ling H, Asi YT et al (2015). Distinct clinical and neuropathological features of G51D SNCA mutation cases compared with SNCA duplication and H50Q mutation. Mol Neurodegener. 27;10:41.
Lücking C and Brice A (2000). Alpha-synuclein and Parkinson’s disease. Cellular and Molecular Life Sciences CMLS 57, 1894–1908
McLaren W, Gil L, Hunt SE. et al (2016). The Ensembl Variant Effect Predictor. Genome Biol 17, 122.
Polymeropoulos,M. H. et al.(1997). Mutation in the α-synuclein gene identified in families with Parkinson’s disease. science 276, 2045–2047.
Recasens, A. & Dehay, B.(2014). Alpha-synuclein spreading in Parkinson’s disease. Front Neuroanat 8, doi: 10.3389/fnana..00159.
Sherry ST, Ward MH, Kholodov M, et al (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res.29(1):308-311.
Siddiqui Irum, Pervaiz Nashaiman and Abbasi Amir. (2016). The Parkinson Disease gene SNCA: Evolutionary and structural insights with pathological implication. Scientific Reports. 6. 24475. 10.1038/srep24475.