Department of Biotechnology, Graphic Era University, Dehradun, Uttarakhand, India.
Corresponding author email: nishantrai1@gmail.com
Article Publishing History
Received: 18/09/2021
Accepted After Revision: 25/12/2021
Non- albicans Candida (NAC) species are responsible for 35-65% of all candidaemias in the general population and are associated with a high rate of morbidity and mortality (about 15% to 35%). The availability of few commercially used antifungal drugs against candidiasis and rapid emergences of antibiotic resistance among NAC species has significantly contributed to their increased global outbreak. Green tea is known for its multi-beneficial effects including antimicrobial potential against Candida. The present study investigated the molecular drug targets of green tea phytocompounds against inhibition of ergosterol biosynthesis in Candida glabrata using in silico tools.The molecular interaction was studied between ligands and essential proteins participating in ergosterol biosynthesis in C. glabrata using autodockvina software. The protein validation and homology modeling estimation were determined by the SWISS MODEL workspace.
The Drug likeness study of all the test ligands was performed using SwissADME, while the toxicity of test compounds was analyzed using the admetSAR 2.0 version.The in silico analyses identified Rutin, Chlorogenic acid, Coumaroylquinic acid, Quercetin, Epigallocatechingallate as the potent phytocompounds with significant molecular binding with Erg 6, Erg 27, Erg 8, Erg 7, Erg 24 respectively. The ADMET data suggested an absence of the CYP2 inhibitors indicating the metabolism of all the tested drug candidates in the intestine and liver.The present study highlighted the possible drug targets of green tea phytocompounds against ergosterol biosynthesis protein in C. glabrata. It is pertinent that the current study has provided preliminary breakthroughs which could lead to exploring their avenues in potent drug development against NAC species. .
Autodockvina, Candida, EGCG, Ergosterol, Rutin
Sirari P, Anand J, Devvret, Thapliya A, Rai N. A Computational Approach to Predict the Molecular Drug Targets Against Candida glabrata. Biosc.Biotech.Res.Comm. 2021;14(4).
Sirari P, Anand J, Devvret, Thapliya A, Rai N. A Computational Approach to Predict the Molecular Drug Targets Against Candida glabrata. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/3dBThbO“>https://bit.ly/3dBThbO</a>
Copyright © Sirari et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Over the last decade, the fungal infections associated with the non-albicans Candida specieshave become a global cause of concern for health professionals (Sardi et al. 2013). Candidiasis is a common fungal infection caused by yeasts from the genus Candida. The disease is associated with a diverse range of infections including mucosal, cutaneous, subcutaneous, and systemic mycoses. Candidiasis which was once considered to be majorly caused by C. albicans, other non- albicans Candida (NAC) species have been recognised to beprimary pathogens associated with increase incidences of Candidiasis.
Epidemiological studies have shown that NAC species are responsible for 35-65% of all candidaemias in the general population (Presterl et al. 2007; Arendrup, 2014; Ahmed et al. 2020).The most common NAC species include C. parapsilosis (20-40% of all Candida species), C. tropicalis (10-30%), C. krusei (10-35%), C. glabrata (5-40%), C. auris (5%), C. lusitaniae (2-8%), and C. guilliermondii (1-5%). Other rare NAC species, are C. rugosa, C. kefyr, C. stellatoidea, C. norvegensisand C. famata (≤1% of Candida infection) (Du et al. 2020).
These NAC species are also identified to be associated with a high rate of morbidity and mortality (about 15% to 35%) particularly in individuals who are immunocompromised like organ transplant recipients, HIV patients, and patients under chemotherapy (Krcmery and Barnes 2002;Bitar 2014; Rudramurthy et al. 2017; Du et al. 2020).A progressive shift in NAC species infection has been linked with indiscriminate use of antibiotics, severe immunosuppressants in immune-compromised such as HIV, cancer or organ transplant patients.The studies have reported that several NAC spp. are inherently resistant to common antifungal drugs and have demonstrated acquired/cross-resistance against antifungals like triazoles (Du et al. 2020).
Indeed, with the availability of a few antifungal drugs commercially used in the treatment of candidiasis and rapid emergences of toxic side effects as well as antibiotic resistance among NAC species, the increased outbreak of NAC associated fungal infection are becoming a global threat (Magill et al. 2006; Pfaller and Diekema 2007; Oberoi et al. 2012; Deorukhkar et al. 2014; Chowdhary et al. 2014; Du et al. 2020).Green tea known for its multi-factorial health properties has been scientifically recognised as a possible antifungal drug representative that could along with standard chemotherapeutics enhance the efficacy of the general treatment approach.
There is plethora of scientific evidence highlighting the antifungal potential of green tea catechins against Candida albicans and identified potent drug candidate’s targeting key enzymes participating in ergosterol biosynthesis (Hirasawa and Takada 2004; Anand et al. 2015; Musial et al. 2020; Huang et al. 2020). As an urgent need to explore alternative potent drug candidates for the development of novel medication against NAC infection, the present study attempts to investigate the inhibitory potential ofgreen tea phytocompounds against ergosterol biosynthesis in C. glabrata using in silicotools.
MATERIAL AND METHODS
In the present in silico study, we selected 15 phytocompounds present in green tea based on their reported antimicrobial activity against Candida spp. These phytocompouds were used as ligands for docking with the ergosterol biosynthesis proteins. Autodockvina(version is 1.1.2 ) tool was used for assessing the molecular interaction based on binding energies. As a positive control, we used azoles (Fluconazole, Itraconazole, and ketoconazole) and Amphotericin B in molecular docking analysis.The code of SMILES for all the aforementioned green tea phytocompounds and positive controls was procured from the online chemical database PubChem (https://pubchem.ncbi.nlm.nih.gov/) (Cheng et al. 2002; Diana et al. 2014).
The 2-D structures of the test ligands were retrieved by converting the SMILES code (.smiles format) into PDB (.pdb) format using chemical interconversion software Open Babel (v 2.3.1). The Drug Likeness reports of all the test ligands were prepared by submitting individual SMILES code for each drug to SwissADME. The toxicity of test compounds was analyzed using the admetSAR 2.0 version (Cheng et al. 2002; Diana et al. 2014).
The 3-D structure of all the proteins participating in the ergosterol biosynthesis was retrieved in PDB format (Table 3) using the PHYRE program (Kelly 2009). The amino acid sequences of ERG proteins in FASTA format were obtained from NCBI [https://www.ncbi.nlm.nih.gov/]. The 3D-protein homology models were generated from the SWISS-MODEL workspace.The protein homology models were validated through a Ramachandran plot created in the SWISS-MODEL workspace.The Molecular interactions were studied for the ERG proteins and the selected phytocompounds as well as positive controls using Autodockvina software. The autodocking tool generated the binding energies of molecular interaction between all the green tea phytocompounds and respective PDB proteins of ergosterol biosynthesis (Waterhouse et al. 2018).
RESULTS AND DISCUSSION
In silico docking analysis: The in silico analysis demonstrated significant molecular interaction between ligands (Tables 1) Positive controls (Table 2) and the docked ERG proteins(Table 3). Among the 15 phytocompounds tested, Rutin and ERG 6 showed the most stable molecular interaction with the least binding energy of -10.80 kcal, compared to the positive control antifungal drugs. It is considered that least the docking energy, the higher will be the binding affinity of a compound with the target protein. Hence, Rutin demonstrated the best interaction among all the investigated green tea phytocompounds.
Among other ligands, Chlorogenic acid, Coumaroylquinic acid, quercetin, Epigallocatechin gallate(EGCG) showed favorable molecular interaction with identified drug targets viz., Erg 6, Erg 27, Erg 8, Erg 7, Erg 24, respectively in ergosterol biosynthesis in C. glabrata. These findings are in agreement with our previous in silico investigation in which we highlighted the inhibitory effect of green tea phytocompounds like chlorogenic acid, EGCG, kaempferitrin against Candida albicans (Anand et al. 2015; Anand et al. 2021).
Table 1 a. Green tea Phytocompounds used as ligands
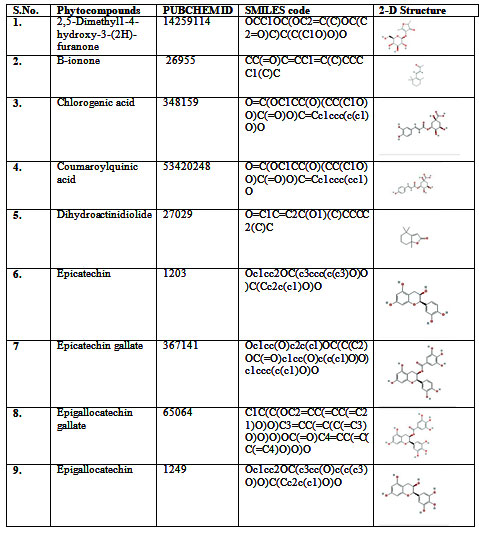
Table 1 b. Green tea Phytocompounds used as ligands
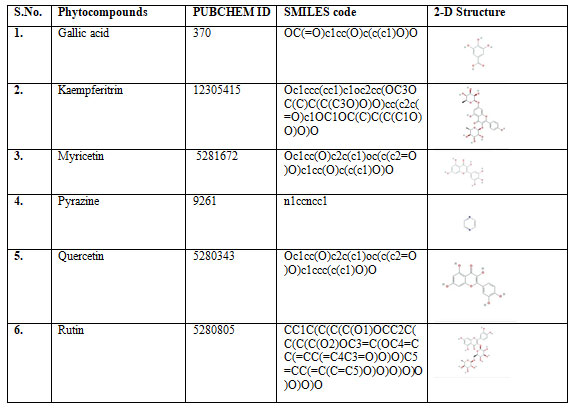
Table 2. Positive controls used in molecular docking study
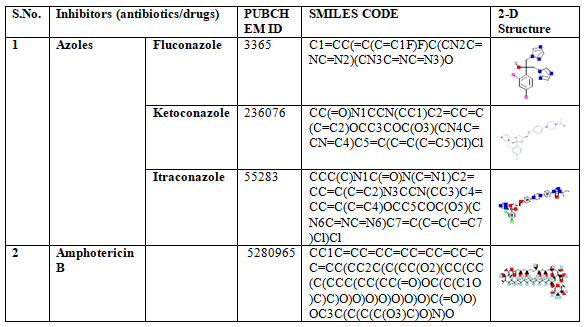
Table 3. Enzymes involved in ergosterol biosynthesis in Candida glabrata
| S.No. | Gene | Protein | Amino acid Sequence
Length |
| 1. | ERG1 | Squalene epoxidase (Erg1) | 206 |
| 2. | ERG2 | C-8 sterol isomerise (Erg2) | 224 |
| 3. | ERG3 | C-5 sterol desaturase (Erg3) | 364 |
| 4. | ERG4 | C-24(28) sterol reductase (Erg4) | 468 |
| 5. | ERG 5 | C-22 sterol denaturase (Erg5) | 364 |
| 6. | ERG6 | Sterol 24-C methyltransferase (Erg6) | 372 |
| 7. | ERG7 | Lanosterol synthase (Erg7) | 733 |
| 8. | ERG8 | Phosphomevalonate kinase (Erg8) | 445 |
| 9. | ERG9 | Squalene synthase (Erg 9) | 443 |
| 10. | ERG 10 | Acetyl coA C-acetyltransferase (Erg10) | 398 |
| 11. | ERG11 | Lanosterol 14-α- demethylase (Erg11) | 533 |
| 12. | ERG12 | Mevalonate kinase (Erg12) | 430 |
| 13. | ERG 13 | 3-Hydroxy-3-methylglutaryl-
coA (HMG-coA) synthase |
281 |
| 14. | ERG20 | Farnesyl pyrophosphate
Synthetase (Erg20) |
351 |
| 15. | ERG 24 | Sterol C-14 reducaase (Erg24) | 437 |
| 16. | ERG25 | C-4 methyl sterol oxidase (Erg25) | 308 |
| 17. | ERG26 | C-3 Sterol dehydogenase (Erg26) | 350 |
| 18. | ERG27 | 3- keto sterol reductase (Erg27) | 348 |
The Ramachandran plot depicted structural stability and showed confirmation of residues in the favorable region (Figure 1; Table 4). A Ramachandran phi-psi plot for all ERG proteins revealed 82.49% to 100.00% of residues are in the allowed region (light gray), and only 0.00% to 4.44% lay in the disallowed region (white). The above analysis of the predicted structure provides supporting evidence that the predicted 3D structure of ERG protein is of good quality.The protein models of ERG proteins showed local similarity to the crystal structures of target templates (Table 4). The Q mean value of protein models was reliable as depicted in the estimated Figure 2. Based on the binding scores, rutin interacted with ERG 6 at the amino acid residues TRY-84, SER-95, GLY-251, ASN-284, TRY-288, ARG-314, PHE-318, and LEU-344 with the least binding energy of -10.80 kcal (Figure 3, Table 5, Table 6, Table 7) (Gao et al. 2020; Anand et al. 2021).
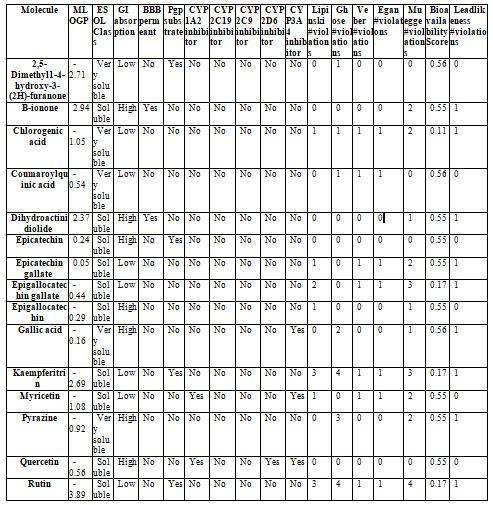
Mammalian metabolism and toxicity of all the proposed green tea phytocompounds as potent antifungal drug agents were tested through ADMET analysis. The ADMET analysis depicted that out of 15 ligands, 13 compounds are non-toxic and obeyed Lipinski’s rule of five. The drug likeliness report determined that Kaempferitrin, rutin, and EGCG are toxic as they have violated two to three rules from Lipinski’s rule of five. Their toxicity level could restrict its application as a potent candidate for antifungal agents; however, before their therapeutic application, the dose regime of these drugs is necessary to be considered and analyzed for the drug toxicity.
The solubility, dissolution, and GI permeability of any drug are essential parameters to be determined before their associated medical effect can be induced. These factors are rate-limiting steps for pre-formulation interpretations in drug development (Shekhawat PB and Pokharkar, 2017; Mitra et al. 2021). In our study, Dihydroactinidiolide and B-ionone, epicatechin, Epigallocatechin, gallic acid, pyrazine, and Quercetin demonstrated high GI absorption, while other ligands were observed to be considerably soluble (Gao et al. 2020; Anand et al. 2021).
Concerning drug potency, the ability of drugs to cross blood-brain barriers (BBB) is estimated. The present study estimated and indicated poor membrane permeation properties of all the tested 13 ligands except Dihydroactinidiolide and B-ionone (Table 8). CYP2 are the family of drug-metabolizing subsets that are involved in the biotransformation of drugs, xenobiotics). The ADMET data suggested an absence of the CYP2 inhibitors indicating metabolism of all drug candidates in the intestine and liver (Table 8) (Tran 2011; Palleri et al. 2013).
The Bioavailability score which is an integral part of the pharmacokinetics paradigm was evaluated less than a standard score of 0.55 for chlorogenic acid, EGCG, kaempferitrin, and rutin and thus depicted their poor bioavailability (Martin 2005). The co-existence of CYP3A4 and Pgp at the same site acts synergistically to reduce the bioavailability of the drug (Mohamed and El-Kadi 2012). Recent studies have also reported poorbioavalibility ofRutin, EGCG, however novel identified transporter system in human can regulate the increase the bioavailibilty of EGCG (Ishii et al. 2019; Fatima et al. 2020).
Table 4. Protein validation and homology modeling estimation by SWISS MODEL workspace.
| S.No. | Gene | Template | Sequence identity (%) | Ramachandran favoured regions (%) | Ramachandran outliers (%) | Q- mean score | Mol probity score |
| 1. | ERG1 | 6c6n.1.A
|
39.30 | 94.09 | 1.48 | -2.39 | 2.04 |
| 2. | ERG2 | 5hk1.1.A | 30.54 | 94.13 | 1.11 | -3.49 | 1.85 |
| 3. | ERG3 | 6sny.1.A | 31.03 | 100 | 0.00 | 0.05 | 0.50 |
| 4. | ERG4 | 4fgs.1.A | 21.49 | 85.98 | 4.44 | -6.29 | 2.32 |
| 5. | ERG6 | 3mgg.1.A | 18.22 | 92.52 | 0.79 | -3.87 | 1.96 |
| 6. | ERG7 | 1w6k.1.A | 39.61 | 93.64 | 1.11 | -2.03 | 1.76 |
| 7. | ERG8 | 3k17.1.A | 20.17 | 82.49 | 5.03 | -7.5 | 2.49 |
| 8. | ERG 9 | 3vj9.1.A | 48.65 | 97.37 | 0.58 | -2.37 | 1.20 |
| 9. | ERG 10 | 5xyj.1.a | 81.11 | 97.72 | 0.25 | 0.81 | 1.17 |
| 10. | ERG11 | 5jlc.1.A | 100 | 96.48 | 0.20 | -0.34 | 1.24 |
| 11. | ERG12 | 2r3v.1.A | 36.67 | 93.32 | 1.87 | -2.29 | 2.39 |
| 12. | ERG 13 | 2p8u.1.A | 46.77 | 91.79 | 0.95 | -4.13 | 2.09 |
| 13. | ERG20 | 4dem.1.A | 46.33 | 96.37 | 0.29 | -0.59 | 1.49 |
| 14. | ERG 24 | 4quv.2.A | 36.74 | 92.86 | 2.62 | -4.97 | 2.26 |
| 15. | ERG25 | 1rh5.1.A | 26.09 | 97.44 | 0.00 | -0.09 | 1.52 |
| 16. | ERG26 | 3lu1.1.A | 20.45 | 90.61 | 1.88 | -3.94 | 2.43 |
| 17. | ERG27 | 4quv.2.A | 31.26 | 90.40 | 3.51 | -5.63 | 1.98 |
Table 5. Binding energies of molecular interaction between ligands and ERG proteins
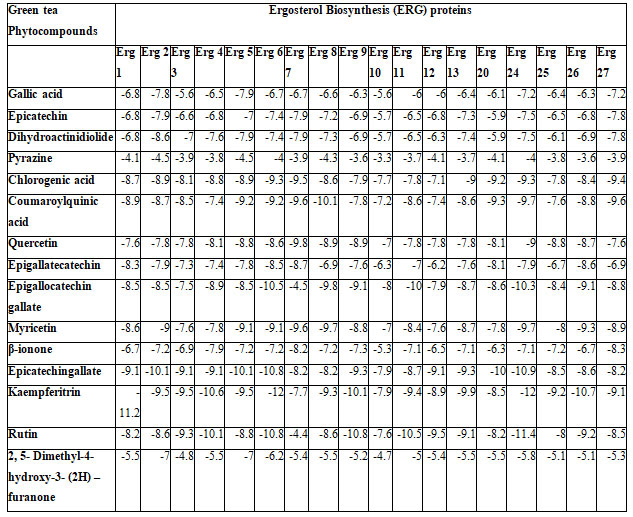
Table 6. Binding energies of molecular interaction of positive controls and ERG proteins
| Inhibitors (antibiotics/drugs) | Targetreceptor | Binding energy | |
| Azoles | Fluconazole | ERG11 | -7.62 |
| ERG 5 | -8.24 | ||
| Ketoconazole | ERG 5 | -11.93 | |
| Itraconazole | -11.69 | ||
| Amphotericin B | ERG | -12.97 | |
Table 7. Most favorable docking poses for the molecular interaction between green tea ligands and ERG proteins.
| S.No. | Most significant molecular bindings | Most favourable docking poses |
| 1 | ERG 27 and Chlorogenic acid | ALA 108, PHE 244, ASN 18, SER 17,GLY 14,LYS 48, ARG 44 |
| 2 | ERG 6 and Rutin | TRY 84, SER 95, GLY 251, LEU 344, ASN 284, TRY 288,PHE 318, ARG 314 |
| 3 | ERG8 and Coumaroylquinic acid | ALA151, SER 150, LYS 191, ALA 377, GLY 148,SER 149, THR 145, LEU 147, GLY 146, LYS 144, PHE 84, PRO 269 |
| 4 | ERG 7 and Quercetin | TYR 91, HIS 226, GLY 377, TRP 583, TYR 703, PHE 695 |
| 5 | ERG 24 and EGCG | ASP 136, GLU 145, TRP 205, TRP 233, MET 230 |
Figure 1
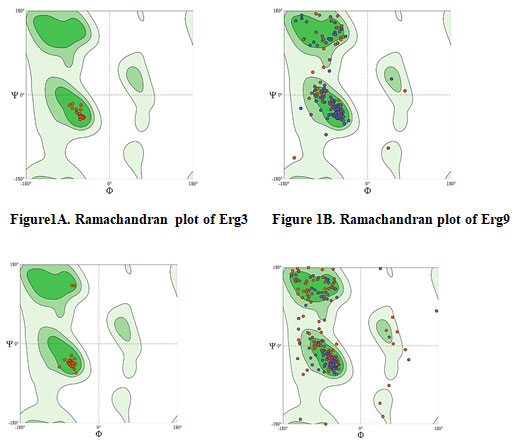
Figure 2: Q mean Z-score value of ERG proteins
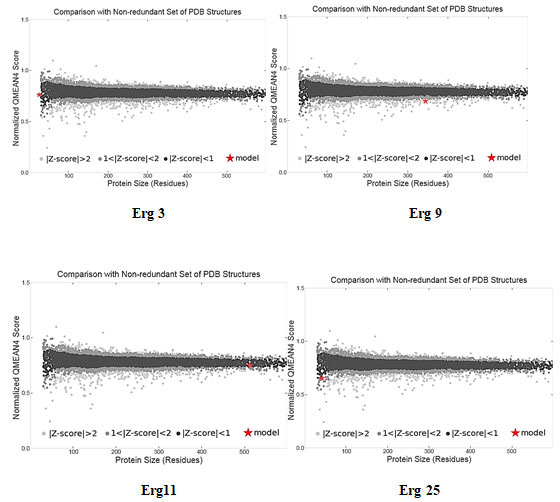
Figure 3: Molecular Docking images of interaction via in silico study.
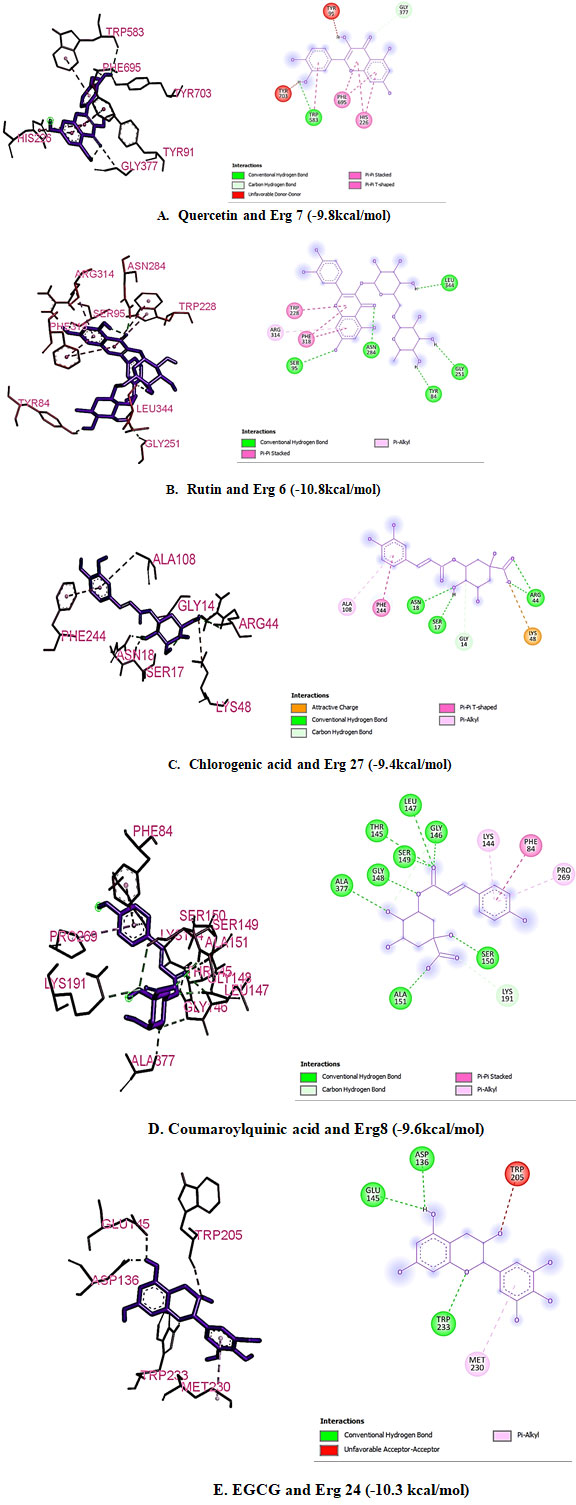
Table 8. Drug likeliness report of green tea phytocompounds.
CONCLUSION
The findings of the present study highlights the antifungal drug targets of green tea phytocompounds against inhibition of ergosterol biosynthesis in Candida glabrata. Rutin, Chlorogenic acid, Coumaroylquinic acid, quercetin, EGCG were screened as the most active green tea phytocompounds with Erg 6, Erg 7, Erg 8, Erg 24, and Erg 27 in ergosterol biosynthesis pathway as their possible drug targets. The poor bioavailability of rutin, chlorogenic acid, and EGCG is a limiting factor and needs further investigation to enhance their bioavailability. The current study is a preliminary analysis that needs to test in vitro to explore the future avenues of potent drug development against NAC species.
ACKNOWLEDGEMENTS
The study was technically supported by Prof. (Dr.) Kamal Ghanshal, Chairman, Graphic Era Deemed to be University, Dehradun.
Conflict of Interests: Authors declare noconflicts of interests to disclose.
REFERENCES
Ahmed S, Shahid M, Fatima N, et al. (2020). Candidemia – Changing trends from Candida albicans to non-albicans Candida from a tertiary care center in western UP, India. CHRISMED Journal of Health and Research Vol 7 Pages 167-72. DOI:10.4103/cjhr.cjhr_12_20
Anand J, Semwal P, Gautam P, et al. (2015). Prediction of novel drug targets in ergosterol biosynthesis pathway: A proposed mechanism of anticandidal activity of green tea phytocompounds Journal of Chemical and Pharmaceutical Research Vol 7 No 2 pages 672-684.
Arendrup MC (2013). Candida and candidaemia. Susceptibility and epidemiology Danish Medical Journal Vol 60 No 11 page B4698.
Bitar B, Khalaf RA, Harastani H et al. (2014). Identification, Typing, Antifungal Resistance Profile, and Biofilm Formation of Candida albicans Isolates from Lebanese Hospital Patients BioMed Research International Vol No 931372 Pages 1-11 DOI:10.1155/2014/931372
Cheng F, Li W, Zhou Y et al. (2012). admetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties Journal of Chemical Information and Modeling Vol 52 No 11 DOI: 10.1021/ci300367a
Chowdhary A, Anil Kumar V, Sharma C et al. (2014). Multidrug-resistant endemic clonal strain of Candida auris in India European Journal of Clinical Microbiology and Infectious Diseases Vol 33 No 6 pages 919–926 DOI: 10.1007/s10096-013-2027-1
Daina A, Michielin O and Zoete V (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules Scientific reports Vol 7 Page 42717 DOI:10.1038/srep42717
Deorukhkar SC, Saini S and Mathew S (2014). Non-albicans Candida Infection: An Emerging Threat Interdisciplinary perspectives on infectious diseases Vol 2014 page 615958 DOIi: 10.1155/2014/615958
Du H, Bing J, Hu T, et al. (2020). Candida auris: Epidemiology, biology, antifungal resistance, and virulence PLoS Pathogens Vol 16 No 10 Page e1008921 DOI:10.1371/journal.ppat.1008921
Fatima S, Gupta P, Sharma S, et al. (2020). ADMET profiling of geographically diverse phytochemical using chemoinformatic tools Future Medicinal Chemistry Vol 12 No 1 Pages 69–87 DOI:10.4155/fmc-2019-0206
Gao C, Cai Y, Zhang K et al. (2020). Association of hypertension and antihypertensive treatment with COVID-19 mortality: a retrospective observational study European Heart Journal Vol. 41 No 22 Pages 2058–2066 DOI: 10.1093/eurheartj/ehaa433
Hirasawa M andTakada M (2004). Multiple effects of green tea catechin on the antifungal activity of antimycotics against Candida albicans The Journal of Antimicrobial Chemotherapy Vol 53 No 2 Pages 225-229 DOI: 10.1093/jac/dkh046
Huang J, Yu H, Hong G, et al. (2020). Antifungal effect of tea extracts on Candida albicans Dental Materials Journal Vol 39 No 4 Pages 664-666 DOI:10.4012/dmj.2019-014
Ishii S, Kitazawa H, Mori T, et al. (2019). Identifcation of the Catechin Uptake Transporter Responsible for Intestinal Absorption of Epigallocatechin Gallate in Mice Scientific Reports Vol 9 No 11014 Pages 1-10 DOI: 10.1038/s41598-019-47214-4
Kelley LA, Mezulis S, Yates CM et al. (2015). The Phyre2 web portal for protein modeling, prediction and analysis Nature Protocols Vol 10 No 6 Pages 854-858 DOI: 10.1038/nprot.2015.053
Krcmery V and Barnes AJ (2002). Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance Journal of Hospital Infection Vol 50 No 4 Pages 243-60 DOI: 10.1053/jhin.2001.1151.
Magill SS, Shields C, Sears CL, et al. (2006). Triazole cross-resistance among Candida spp.: Case report, occurrence among bloodstream isolates, and implications for antifungal therapy. Journal of Clinical Microbiology Vol 44 No 2 Pages 529-35 DOI: 10.1128/JCM.44.2.529-535.2006.
Martin YC (2005). A bioavailability score Journal of Medical Chemistry Vol 48 No 9 Pages 3164-3170 DOI: 10.1021/jm0492002
Mitra D, Verma D, Mahakur M, et al. (2021). Molecular docking and simulation studies of natural compounds of Vitex negundo L. against papain-like protease (PLpro) of SARS CoV-2 (coronavirus) to conquer the pandemic situation in the world Journal of Biomolecular Structure and Dynamics, DOI: 10.1080/07391102.2021.1873185
Mohamed A and El-Kadi AOS (2012). P-glycoprotein effects on drugs pharmacokinetics and drug–drug interactions and their clinical implications. Libyan Journal of Pharmacy and Clinical Pharmacology Vol 1 Page 48154 DOI: 10.5524/LJPCP.v1i0.51150
Musial C, Kuban-Jankowska A and Gorska-Ponikowska M (2020). Beneficial Properties of Green Tea Catechins International Journal of Molecular Sciences Vol 21 No 5 Page 1744. DOIi: 10.3390/ijms21051744.
Oberoi JK, Wattal C, Goel N, et al. (2012). Non-albicans Candida speciesin blood stream infections in a tertiary care hospital at New Delhi, India Indian Journal of Medical Research Vol 136 No 6 Pages 997-1003.
Palleria C, Di Paolo A, Giofre C, et al. (2013). Pharmacokinetic drug–drug interaction and their implication in clinical management. Journal of Research in Medical Sciences Vol18 No 7 Pages 601–610.
Pfaller MA and Diekema DJ (2007). Epidemiology of invasive candidiasis: a persistent public health problem. Clinical Microbiology Review Vol 20 No 1 Pages: 133–163 DOI: 10.1128/CMR.00029-06
Presterl E, Daxbock F, Graninger W et al. (2007). Changing pattern of candidaemia 2001-2006 and use of antifungal therapy at the University Hospital of Vienna, Austria. Clinical Microbiology and Infection Vol 13 Pages 1072–1076 DOI: DOI: 10.1111/j.1469-0691.2007.01812.x
Rudramurthy SM, Chakrabarti A, Paul RA et al (2017). Candida auriscandidaemia in Indian ICUs: analysis of risk factors Journal of Antimicrobial and Chemotherapy. Vol 72 No 6 Pages 1794–1801 DOI: 10.1093/jac/dkx034.
Sardi JCO, Scorzoni L, Bernardi T, et al. (2013). Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options Journal of Medical Microbiology Vol 62 No 1 Pages :10–24 DOI: 10.1099/jmm.0.045054-0
Shekhawat PB and Pokharkar VB (2017). Understanding peroralabsorption: regulatory aspects and contemporary approaches to tackling solubility and permeability hurdles. ActaPharmaceutica Sinica B Vol7 No 3 Pages 260–280 DOI: 10.1016/j.apsb.2016.09.005
Tran N (2011). Blood–brain barrier. In: Encyclopedia of Clinical Neuropsychology. Kreutzer JS, DeLuca J, Caplan B (Eds). Springer, NY, USA.
Waterhouse A, Bertoni M and Bienert S et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes Nucleic Acids Research Vol 46 No W1 pages W296-W303 DOI: 10.1093/nar/gky427


