1Department of Biochemistry, K. S. Rangasamy College of Arts and Science, KSR Kalvi nagar,
Tiruchengode – 637 215, Namakkal, Tamil Nadu, India.
2Department of Biochemistry, Saurashtra University, Rajkot – 360005, Gujarat, India.
3Department of Biochemistry, College of Basic Science and Humanities, Sardarkrushinagar
Dantiwada Agricultural University, Sardarkrushinagar – 385 506, Gujarat, India
Corresponding author email: gsdspu@gmail.com
Article Publishing History
Received:
Accepted After Revision:
Urinary proteomics is the large scale characterization of the protein content of the urine of any organism which is widely used for the diagnosis of a numerous number of human diseases. Human urine, as a collection of substantial amount of proteins, is a non-invasive and readily available biological sample which can be utilized as an effective diagnostic medium for characterizing and tracking the progress of various diseases. The presence of protein and the protein type present in urine varies depending of the nature of the disorder. Thus the ultimate goal of urinary proteome analysis is to compare the protein profile of the normal individual with diseased, so that the indifferent expression of the protein can be identified, finally leading to the designing and development of an innovative biomarker with a diagnostic value. The present study is aimed at standardizing a protocol for the precipitation of the total urinary protein, which can yield high quality protein in higher concentration. Different methods of protein precipitation involving the TCA, Acetone and Acetonitrile have been employed for extracting the total protein content of the urine.
The proteins extracted from the various procedures were validated quantitatively and qualitatively using colorimetric analysis and SDS PAGE respectively. The results of the validating processes have proved that the precipitation of the urine with Acetone and Acetonitrile precipitation resulted in better recovery and integrity of proteins. The study provides an insight to prove protein precipitation from urine to be a successful alternative for the conventional methods of biomarker identification followed by proteome investigation as a part of disease diagnosis and treatment.
Biomarkers, Protein Precipitation, Urinary Proteins
Arunambiga S, Dave G. S, Joshi J, Goswami N, Raval I, Jogani P. A Comparative Study of Urinary Proteins Precipitation Methods. Biosc.Biotech.Res.Comm. 2020;13(3).
Arunambiga S, Dave G. S, Joshi J, Goswami N, Raval I, Jogani P. A Comparative Study of Urinary Proteins Precipitation Methods. Biosc.Biotech.Res.Comm. 2020;13(3). Available from: https://bit.ly/3h1ew6Y
Copyright © Arunambiga et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
A proteome is a complete set of proteins that is normally expressed by the organism, at a specific time in a specific cell or tissue. The proteome can change with time with respect to changes in the physiological behavior of cells (Anderson et al., 2016;Ghosh et al., 2016). Proteomics is used to characterize the entire protein complement of an individual which deals with the study of expression, interactions and functions of proteins(Aslam et al., 2017).The genomics research has provided information on the protein encoding genes that are expressed by the numerous cell types of the human body. The human proteome research is the next step to be taken forward from the information provided by the genomics research(Uhlen, 2008). The applications of proteome research is applied in understanding the protein expression, protein interactions and post translational modifications (Aslam et al., 2017;Chen, 2017). Thus collection of proteins is called proteome and the study of biological protein collection is called proteomics. The use of proteomics will make people to identify the biological disease markers by comparing the protein patterns of biological fluids between normal healthy and diseased individuals (Sanchez-Juanes and Gonzalez-Buitrago, 2019; Sirolli et al., 2020).
A biomarker is a biological component characteristic to any individual which can be measured and appraised as a useful tool to indicate the normal biological processes, disease conditions, or effect of the drug response during therapeutic process (Strimbu and Tavel, 2010). A good biomarker should be accurate, non-invasive and easily collectable for performing the tests(Selleck et al., 2017). Urine is a potential source of a wide variety of components ranging from the small molecular weight metabolites and peptides to high molecular weight macromolecular complexes like exosomes, small vesicles and cytosolic proteins (Hildonen et al., 2016).
In view of this, urine can be used as an excellent tool for the analysis of biomarkers because of its simple composition, abundant availability and frequent sampling from the same individual(Lin et al., 2018; Ding et al., 2020). The use of urine, as a non-invasive source for protein biomarker isolation and identification, is less complex and better method than compared to the use of plasma or cerebrospinal fluid. This is because any change in the human body can be easily tracked with the help of urine and the detection of protein biomarkers is made simple with the use of urine than compared to plasma (Aljaberi et al., 2019; Hao et al., 2020).
Presently, existing tests can be used to determine either the level of total protein or a single protein in urine, whereas the evolving technology of proteomics facilitate the instantaneous analysis of the multiple urinary protein patterns and their link with the process of diagnosis or the individual’s response to any treatment (Amiri-Dashatan et al., 2018).There are various processes available by which the proteins of urine can effectively be isolated and used for the diagnosis of the diseases. Fractionation of the proteins can be done by employing techniques like chromatography, electrophoresis, gel filtration column, dialysis, centrifugal separation or precipitation(Potts, 1965).
Precipitation methods are the most commonly employed method for separation of proteins from the biological samples over a long period of time. Various solvent systems like acetic acid, acetone, acetonitrile, ammonium sulphate, chloroform, ethanol and methanol are added to urine in varying concentrations to facilitate protein precipitation prior to mass spectrometry analysis. The type and quantity of protein precipitated depends on the choice of solvent concentration and combination of solvent systems (Santa et al., 2016; Sanchez-Juanes and Gonzalez-Buitrago, 2019).
The protein precipitation followed by subsequent methods of quantification and validation are the critical steps in proteome studies. Thus a method which can yield high throughput proteins with high recovery and reproducibility in a short duration of time has to be identified. More over the use of non-invasive biomarker source is highly recommended because invasive methods can result in several clinical complications including bleeding, hypotension, local pain, etc.(Bidin et al., 2019). Hence the present study employs urine as a protein source to evaluate the various methods of precipitation which can be used for efficient precipitation of the urinary proteins in large quantities.
MATERIAL AND METHODS
Sample Collection: About 50 – 100ml of the clean, first urine sample in the morning from 4 males and 4 females healthy volunteers (between 20 to 40years of age) was collected in a sterile polypropylene centrifuge tubesas per the method described earlier(Thongboonkerd et al., 2002).
Precipitation of total proteins from urine:The total proteins of the urine samples were fractionated by different precipitation methods. Total nine methods have been employed, three based on TCA precipitation (Magistroni et al., 2009), four based on Acetone precipitation (Magistroni et al., 2009) and two based on Acetonitrile precipitation (Polson et al., 2003; Sirolli et al., 2020; Aljaberi et al., 2019). All the methods are different from each other on the basis of i) temperature and duration of incubation, ii) duration and speed of centrifugation, iii) number and duration of washing step, iv) type and volume of solvents used.
TCA precipitation: Method 1: 50ml aliquot of urine sample was mixed with 12.5ml of 85% ice-cold TCA and incubated at 4°C overnight in a refrigerator. The mixture was then centrifuged at 8000xg for 20min at 4°C and the pellet was washed with 85% ice-cold acetone. The re-suspended pellet was again centrifuged at 12,000xg for 15min at 4°C and the resultant pellet was air dried, re-suspended in 2mlof PBS (Phosphate buffer saline) &50µl of 8% phenylmethylsulfonyl fluoride (PMSF) (protease inhibitor) followed by storage at -20°C for further analysis.
Method 2: An aliquot of 50ml urine sample, mixed with 6.5ml of 85% ice-cold TCA followed by incubation on ice for 2h and spun at 8,000xg for 20min at 4°C. The pellet was washed with 90% acetone by vortexing and then centrifuged at 12,000xg for 20min at 4°C. The pellet was collected, air dried and re-suspended in 2ml PBS &50µl of 8% PMSF and stored at -20°C.
Method 3: To 50ml of the urine aliquot, 25ml of 85% ice-cold TCA was added, mixed and incubated at 4°C for 10min. The mixture was then centrifuged at 12,000xg for 10min at 4°C and the pellet was re-suspended in12.5ml of 90% acetone. The resultant mixture was spun at 14,000xg for 30min at 4°C and the above step was repeated for three times. The subsequent pellet was air dried, re-suspended in 2ml PBS &50µl of 8% PMSF and stored at -20°C.
Acetone Precipitation:Method 1: An aliquot of 50ml urine sample was mixed with half the volume of 85% ice-cold acetone, mixed well and kept on ice for 2 h. The incubated mixture was centrifuged at 12,000xg for 15min at 4°C and then collected pellet was washed with ice-cold acetone. Again the suspension was centrifuged at 12,000xg for 15min at 4°C and the pellet was collected, air dried and re-suspended in 2ml PBS &50µl of 8% PMSF for storage at -20°C.
Method 2: 50ml aliquot of the urine sample was mixed with thrice the volume of 85% ice-cold acetone and kept at -20°C for 30 min. After incubation, the mixture was centrifuged at 13,000xg for 20min at 4°C and the pellet obtained was washed with ice-cold acetone for 3times followed by centrifugation. The pellet thus obtained was air dried and re-suspended with 2ml PBS &50µl of 8% PMSF before storage at -20°C.
Method 3: To 50ml of the urine sample, twice the volume of 85% ice-cold acetone was added, mixed well and incubated on ice for 2h. The mixture was then spun at 12,000 x g for 30min at 4°C and the pellet collected was air dried. The dried pellet was re-suspended in 2ml PBS &50µl of 8% PMSF before the storage at -20°C.
Method 4: 50ml of the urine sample was taken in a centrifuge tube and equal volume of 85% ice-cold acetone was added, followed by incubation overnight at -20°C. The mixture was centrifuged at 12,000xg for 20min at 4°C. The supernatant was discarded and the pellet obtained was air dried, re-suspended in 2ml PBS &50µl of 8% PMSF. Sample was stored at -20°C until the further use.
Acetonitrile precipitation:Method 1: Four times the volume of ice-cold acetonitrile was added to a 50ml aliquot of urine, followed by incubation at room temperature for 90min. After incubation, the mixture was centrifuged at 12,000xg for 15min at 4°C and the pellet obtained was allowed to air dry. The pellet was then re-suspended in 2ml PBS &50µl of 8% PMSF and stored at -20°C.
Method 2: To 50ml aliquot of the urine sample, equal volume of ice-cold acetonitrile was added and then left on ice undisturbed for 2h. The incubated mixture was then spun at 10,000xg for 15min at 4°C and the pellet collected was air dried. The final pellet was re-suspended in 2ml PBS & 50µl of 8% PMSF followed by storage at -20°C.
Estimation of total protein content
The total protein content of the fractionated urine samples were estimated by the method of Lowry (Lowry et al., 1951). Protein was estimated using standard curve prepared by BSA (Bovine Serum Albumin) in the concentration of 200µg/ml.
Qualitative analysis of urinary proteins using SDS PAGE
The urinary proteins extracted by the precipitation methods were analyzed qualitatively by SDS-PAGE (He, 2011). The protein fractions were subjected to separation through 10% Acrylamide gel. The separated bands were then stained using Coomassie Brilliant Blue (CBB) staining.
RESULTS AND DISCUSSION
The present study takes its root from the interest in clinical proteomics and the subsequent discovery of clinically important biological markers. Thus urinary proteomics is receiving an increasing application in clinical diagnosis and follow up measure to be adopted. Even though urine proteomics gains its importance, there are some specific parameters viz., salt removal from sample, presence of minimal quantity of clinically valuable proteins, timing of sample collection, sample storage conditions and adaptation of an efficient protein sample preparation method, which plays a prominent role (Thongboonkerd et al., 2002;González-Buitrago et al., 2007;Candiano et al., 2010;Rastegari et al., 2011). The current study employed nine protein precipitation methods in order to overcome the choice of variation in solubility, hydrophobicity, size, pI and charge of the precipitated proteins from urine (Thongboonkerd et al., 2002; Sanchez-Juanes and Gonzalez-Buitrago, 2019).
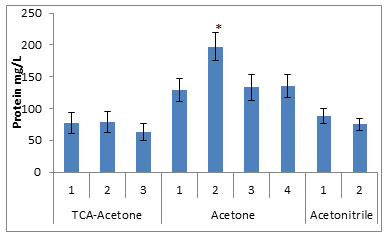
Figure 1: Concentration of urinary proteins extracted by the various precipitation methods.
* compared to all the studied groups, p<0.005
The protein fractions from eight different urine samples precipitated by the nine different methods, when subjected to quantitative analysis by Lowry method, showed highest concentrations (197.3±41.6mg of protein/lit of urine) in the fraction obtained by the method no. 2 of the Acetone precipitation. The other methods of the Acetone precipitation have yielded significantly high amounts of the protein fractions (129.3mg, 133.4mg and 135.3mg of protein/lit of urine) for method 1, 3 and 4 respectively, compared to other studied methods. The TCA precipitation and Acetonitrile precipitation methods yielded low concentration of the proteins, as shown in fig. 1.
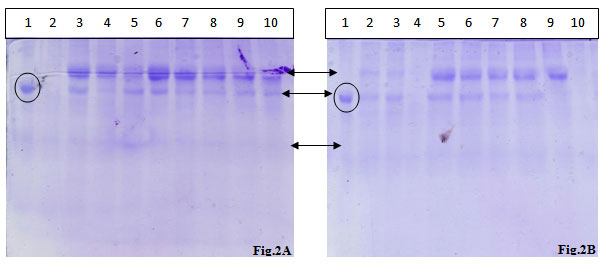
Figure 2: SDS PAGE analysis of urinary proteins extracted from two different samples.
Lane 1: BSA standard,,Lane 2: TCA- Acetone Precipitation Method 1,,Lane 3: TCA- Acetone Precipitation Method 2, Lane 4: TCA- Acetone Precipitation Method 3, Lane 5: Acetone Precipitation Method 1, Lane 6: Acetone Precipitation Method 2, Lane 7: Acetone Precipitation Method 3, Lane 8: Acetone Precipitation Method 4, Lane 9: Acetonitrile Precipitation Method 1, Lane 10: Acetonitrile Precipitation Method 2
The protein fractions of the urinary samples of the eight volunteers were analyzed on the SDS-PAGE and visualized after CBB staining. Almost all the protein fractions displayed the presence of two high molecular weight proteins and one low molecular weight protein in protein fractions.
As shown in fig 2A two high quality bands corresponding to high molecular mass bands were observed with the CBB staining of the gel, in the almost all the wells except TCA-acetone method no. 1. The acetonitrile method 1 yielded two low intensity high molecular mass bands. TCA-acetone method no. 1, acetone methods 1 & 4 and acetonitrile method 2 presented one high intensity band corresponding to a high molecular mass protein.
In fig. 2B, except the TCA-acetone method no. 3 and acetonitrile method no. 2, all the other methods possessed two high intensity high molecular mass spots, out of the two bands, one was completely disappeared in acetonitrile method no. 1. High level of smearing was also observed in all the methods in high intensity, except in acetonitrile method no. 2, where the smearing is of low intensity. Two high intensity spots equivalent to high molecular mass proteins were observed in all the methods; however, one protein band was completely disappeared in all TCA-acetone precipitation methods.The first phase of any clinical proteomics is to find out the most efficient method for optimized purification / separation of the biomarkers from sample.
In this way, for identifying the most effective method of precipitation of the proteins from urine, nine different precipitation methods using TCA-acetone (3 methods), acetone (4 methods) and acetonitrile (2 methods) were selected. All these methods differ from either of them in one or other in the following parameters: i) temperature & duration of incubation, ii) duration & speed of centrifugation, iii) number & duration of washing step and iv) choice & volume of solvents used. The total protein fractionated was quantified using Lowry method and analyzed on SDS-PAGE. Quantitative analysis by Lowry method showed almost similar yield in all the three methods of TCA-acetone precipitation, in case of acetone precipitation, method no. 2 resulted in higher yield of the protein, compared to all other precipitation methods. The other three methods of acetone precipitation have showed significantly high quantity of the proteins. The two methods of acetonitrile have yielded lower quantity compared to acetone methods. However, the highest quantity of the protein yield was found to be in method no. 2 of the acetone precipitation method resulting in good recovery of the urinary proteins.
The qualitative analysis of the proteins from the TCA-acetone method by SDS-PAGE revealed that the bands were either absent or, if present, they are of not intact or of low intensity. The banding patterns of the proteins were varying, in which the bands were sometimes completely absent (Contreras et al., 2008;Walliwalagedara et al., 2010). Besides the presence of some intact bands, the smearing of the bands was observed in almost all the methods. The method 1 of acetonitrile precipitation resulted in better results in terms of band visibility, intensity of the bands and resolution of the bands into intact ones in comparison to TCA methods. However, the band intensity and smearing was similar as in other methods of TCA-acetone precipitation (Walliwalagedara et al., 2010;Contreras et al., 2008).
The protein obtained from acetone precipitation method no. 2 resulted in better bands in terms of number & intensity of the bands, resolution andthe extent of smearing. The TCA-acetone precipitation resulted in lower yield and poor resolution of the bands, which may be due to the presence of salt that may interfere with quantitative estimation and electrophoretic separation, and the prolonged exposure to low pH may also result in denaturation (Walliwalagedara et al., 2010), or the proteins which pI value not in the acidic range, because of the addition of TCA, may be lost during the subsequent washing steps (Jiang et al., 2004;Simpson and Beynon, 2010), or may be due to poor solubility of the proteins (Carpentier et al., 2005;Contreras et al., 2008;Walliwalagedara et al., 2010;Rastegari et al., 2011).Sometimes, extended incubation time also resulted in noticeable positive effect on the protein precipitation. However, change or increase in TCA quantity and increase in number of washing steps doesn’t help much in high protein recovery.
While comparing the three precipitation methods, precipitation using acetonitrile resulted in relatively good protein recovery and good separation on the gel, which is in accordance with the results of published earlier (Romitelli et al., 2007). The acetonitrile precipitation methods also resulted in comparably good results, with variation in the number, intensity and smearing of the bands. This banding pattern of acetonitrile may be due to the property of it to remove some high molecular weight proteins (Alpert, 1999). Evaporation of acetonitrile (Sakuma et al., 1987) may influence the yield of obtained results by both the methods. Though, increase in the solvent volume does makes a sense in higher yield and retrieval of greater number of protein bands, which may have lost in the process with lower volume (Alpert, 1999;Romitelli et al., 2007). Earlier, protein precipitation of yeast growth medium and urine were performed by acetone and ethanol, respectively, our results further supports these findings (Nickerson and Doucette 2020; Zou et al., 2019).
Of all the methods, precipitation by acetone generated better results in terms of protein yield, resolution of the bands, number and intensity of the bands,amount and intensity of smearing, which is in support of earlier results too (Jiang et al., 2004;Walliwalagedara et al., 2010). This is due to the property of acetone in removing the impurities like salts, lipids and pigments that are considered as the possible parameters of interference in urine analysis (Rastegari et al., 2011). Previous reports on urinary protein extraction by DTT method, suggest the similar findings for coronary artery disease (Sun et al., 2019) and hepatocellular carcinoma (Zhao et al., 2020) In addition, the pH of urine is acidic and that of acetone is 6.0 and this minimizes the rate of protein denaturation when compared to TCA precipitation (Thongboonkerd et al., 2002; Ding et al., 2020).
From the analysis made during the study, the acetone precipitation method no. 2 and acetonitrile precipitation method no. 1 yielded better urinary proteins in terms of quality and better resolution of the protein bands on the gel. Acetone precipitation method has been utilized for the In-Solution, MStern, Proteomics iST and S-Trap methods of analysis of urinary proteins, which further supports our finding of urinary protein precipitation methods (Ding et al., 2020).
CONCLUSION
In conclusion, present study explains the comparative study of precipitation methods for precipitation of urinary proteins, insight to open the door of biomarker discovery through non invasive sampling method, such as urine. Human urine can be used as a convenient sample because of its diagnostic importance and source for the discovery of disease biomarkers. The present study uses the human urine for the extraction of total protein by employing nine different methods of precipitation. The purpose is to identify the most efficient method to extract high quantity of total proteins from urine, for futuristic study in proteomics.
The study outcome suggests that method 2 of Acetone precipitation and method 1 of Acetonitrile precipitation resulted in significant recovery of proteins and better resolution on the protein bands in SDS-PAGE. Thus with the obtained results, the above mentioned optimized methods can be applied for the extraction and quantification of urinary proteins. This study can provide an insight to open the arena of biomarker discovery through non-invasive sampling method employing urine as the source of protein.
ACKNOWLEDGEMENT
Present research received the financial assistance and infrastructure facility from Department of Biochemistry, Saurashtra University, Rajkot – 360005, Gujarat, India.
REFERENCES
Aljaberi N, Bennett M, Brunner H I & Devarajan P (2019). Proteomic profiling of urine: implications for lupus nephritis. Expert Review of Proteomics.doi:10.1080/14789450.2019.1592681.
Alpert A J(1999). Size Exclusion High-Performance Liquid Chromatography Of Small Solutes. In: Wu, C.-S. (Ed.) Column Handbook For Size Exclusion Chromatography. San Diego: Academic Press.
Amiri-Dashatan N, Koushki M, Abbaszadeh H-A, Rostami-Nejad M & Rezaei-Tavirani M(2018). Proteomics Applications In Health: Biomarker And Drug Discovery And Food Industry. Iranian Journal Of Pharmaceutical Research, 17, 1523-1536.
Anderson J D, Johansson H J, Graham C S, Vesterlund M, Pham M T, Bramlett C S, Montgomery E N, Mellema M S, Bardini R L, Contreras Z, Hoon M, Bauer G, Fink K D, Fury B, Hendrix K J, Chedin F, El-Andaloussi S, Hwang B, MulliganM S, Lehtio J & Nolta J A(2016). Comprehensive Proteomic Analysis Of Mesenchymal Stem Cell Exosomes Reveals Modulation Of Angiogenesis Via Nuclear Factor-Kappab Signaling. Stem Cells, 34, 601-13.
Aslam B, Basit M, Nisar M A, Khurshid M & Rasool M H (2017). Proteomics: Technologies And Their Applications. Journal Of Chromatographic Science, 55, 182-196.
Bidin M Z, Shah A M, Stanslas J & Lim C T S (2019). Blood and urine biomarkers in chronic kidney disease: An update. Clinica Chimica Acta. doi:10.1016/j.cca.2019.04.069.
Candiano G, Santucci L, Petretto A, Bruschi M, Dimuccio V, Urbani A, Bagnasco S & Ghiggeri G M(2010). 2d-Electrophoresis And The Urine Proteome Map: Where Do We Stand? J Proteomics, 73, 829-44.
Carpentier S C, Witters E, Laukens K, Deckers P, Swennen R& Panis B(2005). Preparation Of Protein Extracts From Recalcitrant Plant Tissues: An Evaluation Of Different Methods For Two-Dimensional Gel Electrophoresis Analysis. Proteomics, 5, 2497-507.
Chen B, Lam TC, Liu L and To C(2017). Post-Translational Modifications And Their Applications In Eye Research (Review). Molecular Medicine Reports. Molecular Medicine Reports, 15, 3923-3935.
Contreras L, Ritter A, Dennett G, Boehmwald F, Guitton N, Pineau C, Moenne A, Potin P & Correa J A(2008). Two-Dimensional Gel Electrophoresis Analysis Of Brown Algal Protein Extracts(1). J Phycol, 44, 1315-21.
Ding H, Fazelinia H, Spruce L A, Weiss D A, Zderic S A, & Seeholzer S H (2020). Urine proteomics: Evaluation of different sample preparation workflows for quantitative, reproducible and improved depth of analysis. Journal of Proteome Research. doi:10.1021/acs.jproteome.9b00772.
Ghosh K, De Graff A M R, Sawle L & Dill K A(2016). Role Of Proteome Physical Chemistry In Cell Behavior. The Journal Of Physical Chemistry. B, 120, 9549-9563.
González-Buitrago J M, Ferreira L& Lorenzo I(2007). Urinary Proteomics. Clinica Chimica Acta, 375, 49-56.
Hao X, Guo Z, Sun H, Liu X, Zhang Y, Zhang L, Sun W & Tian Y (2020). Urinary protein biomarkers for pediatric medulloblastoma. Journal of Proteomics, 103832. doi:10.1016/j.jprot.2020.103832
He F(2011). Laemmli-Sds-Page. Bio-Protocol, 1, E80.
Hildonen S, Skarpen E, Halvorsen T G & Reubsaet L(2016). Isolation And Mass Spectrometry Analysis of Urinary Extraexosomal Proteins. Scientific Reports, 6, 36331-36331.
Jiang L, He L& Fountoulakis M(2004). Comparison Of Protein Precipitation Methods For Sample Preparation Prior To Proteomic Analysis. Journal Of Chromatography A, 1023, 317-320.
Lin L, Yu Q, Zheng J, Cai Z & Tian R(2018). Fast Quantitative Urinary Proteomic Profiling Workflow For Biomarker Discovery In Kidney Cancer. 15, 42.
Lowry O H, Rosebrough N J, Farr A L & Randall R J(1951). Protein Measurement With The Folin Phenol Reagent. J Biol Chem, 193, 265-75.
Magistroni R, Ligabue G, Lupo V, Furci L, Leonelli M, Manganelli L, Masellis M, Gatti V, Cavazzini F, Tizzanini W & Albertazzi A (2009). Proteomic Analysis Of Urine From Proteinuric Patients Shows A Proteolitic Activity Directed Against Albumin. Nephrol Dial Transplant, 24, 1672-81.
Mary S, Currie G, Schutte A E, & Delles C (2020). Cardiovascular proteomics. Precision Medicine for Investigators, Practitioners and Providers, 263–270. doi:10.1016/b978-0-12-819178-1.00025-3.
Nickerson J L and Doucette A A. (2020). Rapid and Quantitative Protein Precipitation for Proteome Analysis by Mass Spectrometry. J. Proteome Res. 2020, 19, 2035−2042.
Polson C, Sarkar P, Incledon B, Raguvaran V & Grant R(2003). Optimization Of Protein Precipitation Based Upon Effectiveness Of Protein Removal And Ionization Effect In Liquid Chromatography-Tandem Mass Spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci, 785, 263-75.
Potts A M(1965). Methods For Separation Of Proteins. Investigative Ophthalmology & Visual Science, 4, 531-538.
Rastegari E, Ahmad Z, Spencer D F & Ismai M(2011). Two-Dimensional Profiling Of Proteins From Curculigo Latifolia Fruit By Three Different Extraction Protocols. Journal Of Medicinal Plants Research, 15, 3719-3724.
Romitelli F, Santini S A, Chierici E, Pitocco D, Tavazzi B, Amorini A M, Lazzarino G & Di Stasio E(2007). Comparison Of Nitrite/Nitrate Concentration In Human Plasma And Serum Samples Measured By The Enzymatic Batch Griess Assay, Ion-Pairing Hplc And Ion-Trap Gc-Ms: The Importance Of A Correct Removal Of Proteins In The Griess Assay. J Chromatogr B Analyt Technol Biomed Life Sci, 851, 257-67.
Sakuma R, Nishina T& Kitamura M(1987). Deproteinizing Methods Evaluated For Determination Of Uric Acid In Serum By Reversed-Phase Liquid Chromatography With Ultraviolet Detection. Clin Chem, 33, 1427-30.
Sánchez-Juanes F, & González-Buitrago J M (2019). Sample Treatment for Urine Proteomics. Emerging Sample Treatments in Proteomics, 125–135. doi:10.1007/978-3-030-12298-0_5.
Santa C, Anjo S I, & Manadas B (2016). Protein precipitation of diluted samples in SDS-containing buffer with acetone leads to higher protein recovery and reproducibility in comparison with TCA/acetone approach. Proteomics, 16(13), 1847–1851. doi:10.1002/pmic.201600024.
Selleck M J, Senthil M & Wall N R(2017). Making Meaningful Clinical Use Of Biomarkers. Biomarker Insights, 12, 1177271917715236-1177271917715236.
Simpson D M & Beynon R J(2010). Acetone Precipitation Of Proteins And The Modification Of Peptides. Journal Of Proteome Research, 9, 444-450.
Sirolli V, Pieroni L, Di Liberato L, Urbani A & Bonomini M (2019). Urinary Peptidomic Biomarkers in Kidney Diseases. International Journal of Molecular Sciences, 21(1), 96. doi:10.3390/ijms21010096.
Strimbu K & Tavel J A(2010). What Are Biomarkers? Current Opinion In Hiv And Aids, 5, 463-466.
Sun H, Wang D, Liu D, Guo Z, Shao C, Sun W, Zeng Y (2019). Differential urinary proteins to diagnose coronary heart disease based on iTRAQ quantitative proteomics. Analytical and Bioanalytical Chemistry, https://doi.org/10.1007/s00216-019-01668-7.
Thongboonkerd V, Mcleish K R, Arthur J M & Klein J B(2002). Proteomic Analysis Of Normal Human Urinary Proteins Isolated By Acetone Precipitation Or Ultracentrifugation. Kidney Int, 62, 1461-9.
Uhlen M(2008). A New Era For Proteomics Research? Genome Biology, 9, 325.
Walliwalagedara C, Kerulen H V, Right T C & Wei R(2010). Comparison Of Sample Preparation Methods For The Resolution Of Metal-Regulated Proteins In Helianthus Annuus By 2-D Gel-Electrophoresis. The Open Proteomics Journal, 3, 20-25.
Zhao Y, Lib Y, Liu W, Xing S, Wang D, Chen J, Sun L, Mu J, Liu W, Xing B, Sun B, and He F (2020). Identification of noninvasive diagnostic biomarkers for hepatocellular carcinoma by urinary proteomics. Journal of Proteomics, 225, 1-9.
Zou L, Wang X, Guo Z, Sun H, Shao C, Yang Y and Sun W (2019). Differential urinary proteomics analysis of myocardial infarction using iTRAQ quantification. Molecular Medicine Reports 19, 3972-3988.