1,2Post Graduate Department of Biosciences and Biotechnology, Fakir Mohan University, New campus, Nuapadhi, Balasore, Odisha-756020, India
3School of Life Sciences, Hyderabad Central University, Prof C. R. Rao Road, P.O. Central University, Gachibowli, Hyderabad, Telangana 500046, India
Corresponding author Email: shubhashreemahalik@gmail.com
Article Publishing History
Received: 21/10/2018
Accepted After Revision: 21/11/2018
Microbes are rich sources of natural products like secondary metabolites, enzymes and proteins. In this context the primary objective of the present work is to isolate microbes from natural habitats and characterize them on the basis of their ability to produce metabolites. Since estuaries are the junction of marine and coastal habitats and harbour a plethora of microbes therefore in this study estuaries along the coastal district of Balasore, Odisha were screened for cellulase secreting bacteria. Cellulase forms an important component of enzyme cocktail used for degradation of lignocellulosic biomass for production of biofuel. Several rounds of sampling, pure culture, morphological, biochemical and phylogenetic screening led to the identification of Lysinibacillus sp. having the ability to secrete cellulase. Physical as well as nutritional characterization like optimization of media, temperature, carbon and nitrogen requirements was performed to enhance the biomass formation. The isolated strain of Lysinibacillus sp. showed higher biomass and growth rate at 37°C, in Terrific Broth media supplemented with 0.5% Glucose and 0.5% Sodium Nitrite. Submerged fermentation under anaerobic condition at 37°C for 5 days led to release of 9.85µmole of glucose/ml of enzyme.
Lysinibacillus, Estuary, Cellulase, Phylogenetic Analysis
Mahalik S, Mohapatra D, Kumar D. Cellulase Production in Lysinibacillus Sp Isolated from the Estuaries of Odisha. Biosc.Biotech.Res.Comm. 2018;11(4).
Mahalik S, Mohapatra D, Kumar D. Cellulase Production in Lysinibacillus Sp Isolated from the Estuaries of Odisha. Biosc.Biotech.Res.Comm. 2018;11(4). Available from: https://bit.ly/2m1WwSa
Introduction
Several microorganisms are considered to be extremophiles as they are adapted to extreme environmental conditions such as high or low temperature, alkaline or acidic water, high pressure and substrate limitations. These characters make them potential industrial organisms as they produce several interesting metabolites to cope these extreme conditions. Various literatures are available where microorganisms isolated from extreme geographical locations have been used for production of hormones, proteins, enzymes and other primary and secondary metabolites (Coker, 2016; L Bergquist et al., 2014; Littlechild, 2015; Poli et al., 2017; Stierle & Stierle, 2014; Yin & Chen, 2015). One such extreme environment is the estuary. Estuaries have rich biodiversity due to variation in temperature, pH, salinity and availability of salts and other minerals (Campbell & Kirchman, 2013; Lallias et al., 2015). Several Bacillus species are known to be cellulase producers (Irfan et al., 2017; Sanjeev et al., 2017), but very few reports are available on cellulase production by Lysinibacillus strains
These brackish water bodies are an amalgamation of both fresh water from rivers and saline water from tidal waves of sea. This character makes it a very productive habitat for various flora and fauna including microorganisms (Moyle et al., 2010). Odisha is one of the coastal states situated on the eastern part of India having a coast line of 480 km. The coastline of the Balasore district is in the shape of a strip with a length of 81 km and 26 km wide. Several estuarine rivers like Budhabalanga, Subarnarekha flood the coastal areas. Many studies have been carried out on the macro flora and fauna of this area but very few documented information is available regarding the microbial biodiversity, (Subudhi & Patra, 2013; Sujana et al., 2015 Bomble et al., 2017).
Cellulases are a group of enzyme that degrade cellulose by hydrolyzing â-1,4 linkages in cellulose chains. Naturally, the cellulase is produced from widespread sources like fungi, bacteria, protozoans, plants, and animals (symbiotic bacteria in few ruminants and termites). The biotechnological application of lignocelluosic biomass in several industries like paper, textile, food, biofuel as well as agriculture has led to extensive research on production, biochemical as well as enzymatic characterization of cellulase. Microbes from many bacterial genera have proved to be a good source for cellulase production at industrial scale, Juturu & Wu, 2014; Kuhad, Gupta, & Singh, 2011 Kuhad et al., 2016 and Bomble et al., 2017). In the present study an attempt has been made to isolate cellulase producing bacteria from the estuaries around the Balasore district and optimize the culture conditions for the production of
cellulase.
Materials & Methods
Sample collection and pure culture: Soil sediments up to a depth of 6-10 inch were collected from different spots of the Khandia estuary (21°19’1.65”N; 86°53’32.99”E) in Balasore district near the mouth of Khandia river. Soil samples were serially diluted in 1X PBS solution and plated on nutrient agar (NA) plate. The plates were incubated at 37°C for 24 hour and the colonies obtained were re-streaked on fresh NA plate and the mother plate was allowed to incubate further. The colonies that appeared after 48 hour and 72 hour of incubation were also re-streaked on fresh NA plates to obtain isolated colonies. Dilution streaking was performed for all the isolated colonies that were obtained from various samples. This process was repeated for several times till pure cultures were obtained. All pure cultures were labelled and stored at 4°C in NA stabs.
Screening for cellulase producing strains: The isolates were screened on the basis of their ability to secrete cellulase. NA plates were overlayed with 0.5% carboxymethyl cellulose (CMC). CMC acts as substrate for cellulase enzyme. The bacterial cultures were spread on the CMC supplemented plates. Post incubation the plates were stained with 0.1% Congo Red. The plates were destained with 1N NaCl solution and observed for clear zones (Meddeb-Mouelhi, Moisan, & Beauregard, 2014).
Morphological and physiological characterization of the isolates: The shape, size, elevation, margin and colour of the colony were observed and the morphology of the isolates was determined using Grams Staining method. Biochemical test like Citrate Utilization, Triple Sugar Iron, Mannitol Motility, Gelatin Hydrolysis, Oxidase, Indole production and antibiotic resistance tests were performed. Citrate Utilization was performed on Simmon’s citrate agar medium (Himedia, M099), TSI was tested on Triple Sugar-Iron Agar (Himedia, MM021), Mannitol Motility was checked on Mannitol Motility Test Medium (Himedia, M770), Gelatin Hydrolysis was checked on Nutrient gelatin medium prepared in lab. The presence of oxidase enzyme was checked using Oxidase disc (Himedia, DD018) and Indole production was checked using Kovac’s strip (Himedia, DD019). All experiments were performed using standard protocol as recommended by product manual. Resistances for ampicillin, kanamycin, tetracycline, penicillin and streptomycin were checked by disc diffusion method at 50μg/μl, 5μg/μl, 0.5μg/μl and 0.05μg/μl concentration for all the antibiotics.
Growth Characteristics of the Isolates: The isolates were grown in 4 different media (Nutrient Broth, Terrific Broth, Marine Broth and Artificial Sea Water) at 25 ºC and 37ºC. The experiment was performed in a microbioreactor (m2p labs BioLector) where biomass was continuously monitored till the population reached at stationary phase. The growth was also monitored under various carbon sources (Glucose, Maltose, Lactose, Xylose, Cellobiose and starch) and nitrogen sources (Sodium nitrate, Di-ammonium hydrogen citrate, Ammonium nitrite, Tryptone, Yeast extract and Ammonium chloride). The concentration of carbon and nitrogen supplemented to the media were 0.5% each. Similarly salt tolerance was checked for the isolates in TB medium supplemented with various concentration of NaCl ranging from 0.5% to 12%.
Isolation of Genomic DNA: 5ml bacterial culture was inoculated and incubated overnight. After incubation the culture was transferred to a centrifuge tube and centrifuged at 10,000 rpm for 10 minutes until a compact pellet was formed. The supernatant was discarded and the pellet was resuspended in a mixture of 567μl TE buffer and 5μl RNAse A by repeated pipetting. 15μl 10% SDS and 4μl proteinase K (18 mg/mL) was added. It was mixed thoroughly and incubated 15-20 minutes at 65°C until all the cells are lysed.100μl of 5M NaCl was added and mixed thoroughly. 80μl of CTAB/NaCl (10% w/v; 0.7M) solution was added and mixed thoroughly and incubated for 10min at 65°C. Equal volume of chloroform/isoamyl alcohol (0.7-0.8ml) was added and mixed thoroughly and centrifuged for 5min at 10,000 rpm. 1 volume of isopropanol was added to the supernatant, shaken and centrifuged. The pellet obtained was washed with 70% ethanol, dried and dissolved in 50μl TE buffer.
PCR and sequencing: The 16s rRNA gene was amplified from the genomic DNA of all the isolates. The primers used in the study are as follows (Frank et al., 2008; Karakasidou et al., 2018): BAC27F AGAGTTTGATCCTGGCTCAG BAC1492RGGTTACCTTGTTACGACTT: The PCR was carried out at initial denaturation of95°C/5min, denaturation of 95°C/30sec, annealing at 42°C/1min, extension at 72°C/1min 30sec and final extension at 72°C/5min. This was repeated for 30 cycles.The sequencing of the purified PCR product was done using the BAC27F forward primer. The work was outsourced form SciGenom Labs Pvt. Ltd., Kerala, India.
Phylogenetic Analysis: The molecular characterization of all the isolates were done by 16s rRNA sequencing. The phylogenetic was prepared using MEGA 7 program (Sudhir Kumar, Stecher, & Tamura, 2016) with Neighbor Joining method. Sequence of Lysinibacillus sp. KEI3 was BLAST in EZ taxon (Chun et al., 2007) and only validly published sequences were taken as references in tree formation. Boot strap replication was performed 1000 times.
Submerged Fermentation: Bacterial isolates were cultured in Terrific broth medium under submerged fermentation conditions. 50 ml TB medium was prepared in 250 mL flask. The medium was supplemented with 0.5% Glucose and 0.5% Sodium Nitrite. Medium was sterilized by autoclaving. The flasks were incubated in a shaking incubator at 37 ± 2 °C for 5 days and then crude enzyme was extracted by centrifugation at 10,000 rpm for 20 min at 4 °C. The cell free culture filtrate (CFCF) was used as crude enzyme to test Cellulase activity. Cellulase activity was measured by DNS assay.
Results
Isolation of Bacterial Colonies: After serial dilution, plating of soil samples and incubation, more than 50 colonies were obtained. Out of these, 11 pure cultures were obtained which were screened for their ability to degrade CMC by plating them on CMC agar plate and staining with Congo Red. Isolate 3 showed clear zones on CMC agar plates indicating their ability to secrete cellulase. Morphological and biochemical characterization was performed for Isolate 3 and the observations are presented in Table 1. The results indicate that the isolates belong to genus Bacillus.
| Table 1: Biochemical characters of Lysinibacillus sp.KEI-3 | ||||||||||||
| TEST | Gram’s staining | TSI | Mannitol Motility | Citrate utilization | Gelatin Hydrolysis | Oxidase | Indole | Ampicillin | Kanamycin | Tetracyclin | Penicillin | Streptomycin |
| KEI-3 | + | K/A | – | – | + | + | – | + | + | + | + | + |
| (-) represents negative response to the test; (+) represents positive response or susceptibility to the test. | ||||||||||||
Phylogenetic Analysis: Genomic DNA isolated from pure cultures were subjected to PCR amplification of the 16s RNA using BAC27F and BAC1498R primers which produced around 1450bp long 16S rRNA gene. Phylogenetic tree was prepared using MEGA 7 program and Isolate 3 was identified to be Lysinibacillus sp. which has maximum similarity to Lysinibacillus fusiformis strain 4 (KF916674) (Figure 1). The Isolate was named as Lysinibacillus sp.KEI-3.
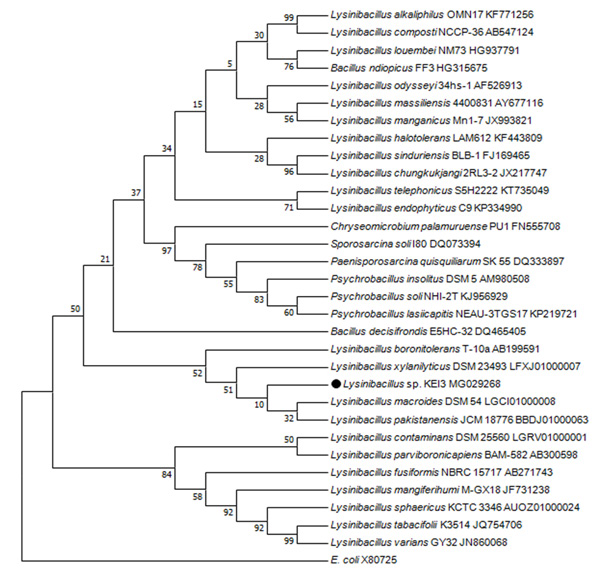 |
Figure 1: Phylogenetic tree prepared using MEGA 7 software with Neighbor Joining methodon the basis of 16sRNA sequencing. |
Effect of different media on growth
Lysinibacillus sp. KEI-3 was inoculated in Nutrient Broth (NB), Terrific Broth (TB), Marine Broth (MB) and Artificial Sea Water supplemented with Glucose and Tryptone (SWGT) media and grown at 25°C and 37°C. The growth was monitored for 36 and 24 hour respectively and plotted to calculate specific growth rate. It was observed that when isolates were cultured at 25°C it had a very long lag phase and the specific growth rate was also slow in comparison to cells growing at 37°C (Figure 2). This phenomenon was observed for all media used for the study except TB medium, where irrespective of temperature the isolate Lysinibacillus sp.KEI-3 had a high specific growth rate. For TB medium the specific growth rate was 0.558h-1 at 37°C whereas at 25°C it was 0.451h-1 which is almost comparable. Interestingly in SWGT media the isolate had almost 20 hour long lag phase at 25°C after which there was increase in growth of the cells and the maximum specific growth rate achieved at 25°C was 0.338h-1 whereas at 37°C the it was 0.557h-1 (Figure 3).
 |
Figure 2: Growth profile of Lysinibacillus sp. KEI-3 strain in different media and different temperature. (A) growth curve at 37ºC, (B) growth curve at 25ºC. |
 |
Figure 3: Specific Growth rate of Lysinibacillus sp. KEI-3 strain in different media and different temperature. |
It was interesting to observe that the isolate could not grow well in LB and MB. The reasons for LB are obvious that it is nutritionally less rich and complex than TB due to which the growth rate was slow. MB is high in salt concentration and probably the isolate do not have high salt tolerance, which could be the possible reason for limited growth in MB medium. SWGT medium was formulated in lab where the composition of artificial seawater was almost similar to the original seawater but the salt concentration was less. Further it was enriched by addition of Glucose and Trypotone which are very good sources of carbon and complex nitrogen requirements.
Effect of different carbon source on growth: Six different carbon sources namely Glucose, Lactose, Starch, Xylose, Maltose and Cellobiose were selected in present study. These carbon sources were added at a final concentration of 0.5% separately in TB media. Lysinibacillus sp.KEI-3 was inoculated in these media and grown at 37°C. The growth was measured after 24 hour of incubation. It was observed that the isolate could effectively utilize both pentose and hexose sugars. Biomass profiling revealed that Lysinibacillus sp. KEI-3 effectively utilize monosaccharide (glucose and xylose), disaccharides (maltose, lactose and cellobiose) as well as polysaccharide (starch) (Figure 4).
 |
Figure 4: Growth profile of Lysinibacillus sp. KEI-3 strain with different carbon sources. |
Effect of different nitrogen source on growth: Different bacterial species utilize different nitrogen sources for their growth. The different nitrogen sources selected for this study are sodium nitrate, di-ammonium hydrogen citrate, ammonium nitrate, tryptophan type-1, yeast extract, and ammonium chloride. TB media was prepared to which different nitrogen source was added separately at a final concentration of 0.5%. The isolated strain was inoculated in these media and grown under 37°C. The growth was measured after 24 hour of incubation. It was observed that Lysinibacillus sp. KEI-3 can grow well in both organic as well as inorganic nitrogen sources and the maximum biomass was formed in media supplemented with yeast extract, tryptone and sodium nitrite (Figure 5).
Effect of different salt concentration on growth: To check the salt tolerance capacity, Lysinibacillus sp.KEI-3 was inoculated in TB medium supplemented with various concentration of NaCl. The salt tolerance was checked for concentration range of 0.5-12%. The strain was inoculated and incubated at 37°C for 24 hours. After incubation the biomass was measured spectrophotometrically by reading OD at 600nm.Since isolates could not grow well in Marine Broth (MB) due to its high salt content, therefore supplementing different concentrations of NaCl in the medium was used to check maximum salt tolerance of the Lysinibacillus sp.KEI-3which showed tolerance till 1% and beyond that there was decline in biomass formation. But it could grow till 12% with a lower growth rate (Figure 6).
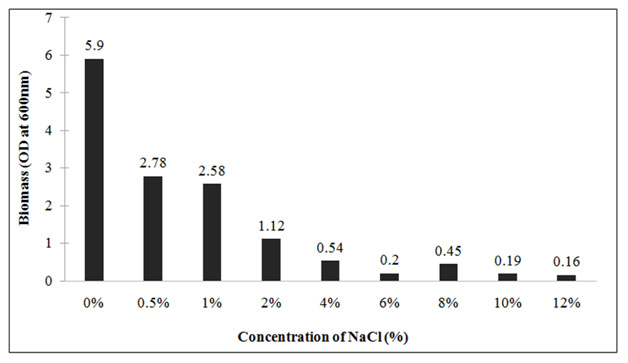 |
Figure 6: Growth profile of Lysinibacillus sp. KEI-3 strain at different salt (NaCl) concentration. |
Submerged fermentation
Lysinibacillus sp.KEI-3 was further subjected to submerged fermentation for production of cellulase. Terrific Broth was supplemented with optimized carbon and nitrogen sources (glucose/sodium nitrite) that showed higher biomass for Lysinibacillus sp.KEI-3. Anaerobic fermentation was continued at 37°C for 4 days. Samples were collected and cellulase produced was quantified by DNS assay. The results are represented in the form of µmole of glucose (reducing equivalent) released per ml of cellulase enzyme produced. Through the time course sampling, it was observed that till 48 hours there was no significant increase in cellulose production. Whereas, after 96 hours of incubation there was 2.5 times increase in cellulose production (Figure.7). This was a promising result and the production could be increased by further scaling up and optimization.
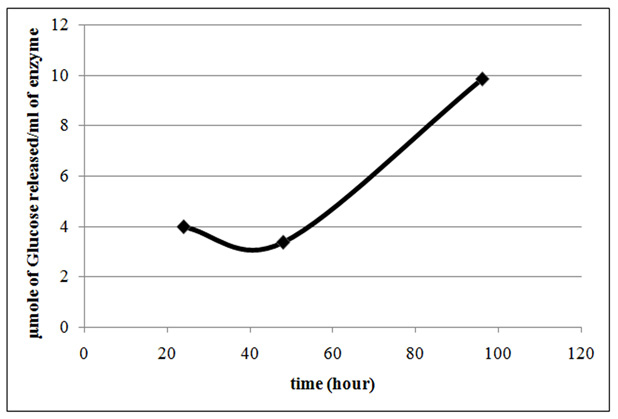 |
Figure 7 : Cellulase production profile of Lysinibacillus sp. KEI-3 strain. Cellulase productivity is represented in the form of µmole of Glucose released per ml of cellulase produced. |
Discussion
70% of the planet’s surface area is covered by oceans. The coastal environments support huge diversity of microbial life. But still only a small fraction of the species has been cultured and identified till date due to culture related problems. Heavy pollution has led to severe destruction of marine biological diversity (Abreo et al., 2015; Baum et al., 2015).Trawler fishing, pollution from industries and drainage system has led to increase in eutrophication which is further leading to change in aquatic ecosystem as well as destruction of habitats. This has led to decline in microbial biodiversity. So it has become imperative to identify and isolate the organisms and make a database, so that the information can be evaluated time to time to check for the loss of species in the marine world. Estuaries are dynamic in nature in terms of the nutritional content and the associated microbial population and this is mainly influenced by the convergence of fresh water and sea. The biochemical environment of the estuaries makes it ideal for availability of diverse microbial communities (Andersson et al., 2014; Zhang et al., 2014). In different parts of the world several studies have been undertaken to describe the microbial diversity along the estuaries and their physiochemical relation with the surrounding environment ( Reed & Martiny, 2013; Sun et al., 2014 and Lallias et al., 2015).
Even though the eastern part of coastal India has several estuaries, no substantial studies have been undertaken to highlight the microbial diversity or the potentialities for bioprospecting. In this work both the aspects were covered, where pure cultures were isolated from soil samples collected from estuaries and screened on the basis of their ability to secrete cellulase enzyme. Morphological and biochemical characterization of the Isolate 3 showed similarity with Bacillus species. Phylogenetic analysis was done using 16s rRNA sequencing and the strain was identified to be Lysinibacillus sp.KEI-3.These soil bacteria are rod shaped and gram positive. Lysinibacillus fusiformis, Lysinibacillus sphaericus, Lysinibacillus boronitolerans, Lysinibacillus macroides and Lysinibacillus xylanilyticus are some of the strains that have been isolated and characterized previously. The strains belonging to this genus have several industrial importance such as xylan degradation (Lee et al., 2010), biodegradation of low-density polyethylene (Esmaeili et al., 2013), biotransformation of Indole to 3-Methylindole (Arora et al., 2015), biological pest control (Rojas-Pinzón & Dussán, 2017).
These characters make this genus an interesting target for microbiological studies.The biomass production was optimized under several physical and chemical parameters such as temperature, media composition, carbon source, nitrogen source and salinity tolerance. The organisms showed high growth rate at 37ºC in TB medium and SWGT medium. Bacterial species can utilize both pentose and hexose sugars (Cook et al., 1993; Kim et al., 2009; Liu et al., 2008). Each species has its own ability to breakdown and utilize several carbon sources. Also the preference of carbon source varies from species to species (Brückner & Titgemeyer, 2002; Görke & Stülke, 2008).Therefore the biomass yield was assessed with various carbon sources which showed that this species have a broad range of carbon preferences. They could well utilize both pentose as well as hexose
sugars.
Organic as well as inorganic nitrogen sources are critical for growth of microorganisms (Wheeler & Kirchman, 1986). The carbon to nitrogen (C/N) ratio is important in a biological process (Cleveland & Liptzin, 2007). Microorganisms require a proper nitrogen supplement for metabolism during fermentation (Lin & Lay, 2004). Therefore the optimal nitrogen was characterized in order to obtain maximum biomass. It was observed that Lysinibacillus sp.KEI-3 could produce higher biomass when the media was supplemented with Sodium Nitrite.Since the Isolates could not grow well in Marine Broth (MB) due to its high salt content, therefore supplementing different concentrations of NaCl in the medium was used to check maximum salt tolerance. Lysinibacillus sp.KEI-3 had higher salt tolerance and it can grow even at 12 % NaCl.
Several Bacillus species are known to be cellulase producers (Irfan et al., 2017; Sanjeev et al., 2017), but very few reports are available on cellulase production by Lysinibacillus strains (Khianngam et al., 2014). Therefore submerged fermentation was done under the above-optimized conditions and it was observed that the cell could accumulate significant amounts of cellulase. Even though the total units of cellulase produced are low as compared to reported species, but it is a positive sign that the isolated strain is a cellulase producer. Further optimization of physical as well as bioprocess parameters could lead to accumulation of higher levels of cellulase at high cell density cultures.
In the present scenario, the carbohydrolytic bacteria or the lignocellulose degrading bacteria have a greater industrial demand given their application in saccharification of lignocellulose for biofuel production. In this context cellulase is the most common enzyme used in the cocktail for degradation of lignocellulosic biomass (Sindhu et al., 2016). Therefore there has been a continuous effort to screen and isolate efficient cellulase producing microbes. These strains are isolated from various geographical areas and also grown under various cheap and alternative substrates for production of enzymes. Apart from characterization of bacteria, process parameters as well as culture conditions are also being optimized to enhance bacterial biomass to increase the yield of cellulase. Optimization of media, carbon, nitrogen, salinity as well as temperature requirement led to increased production of cellulase in the Lysinibacillus sp.KEI-3.This isolated strain could further be screened for other enzymes like xylanase and pectinase, which would make it a potential strain for saccharification of lignocellulose biomass.
Acknowledgment
The authors acknowledge the Bioprocess and Biosystems Engineering lab, JNU, New Delhi for providing all necessary help. Also P.G. Department of Biosciences and Biotechnology, Fakir Mohan University, Balasore, Odisha is acknowledged for providing infrastructure for carrying out the experiments.
Declaration Of Interest Statement
The authors declare that they have no competing interests.
Data Availability Statement
GenBank accession number for the 16s rRNA nucleotide sequence of Lysinibacillus sp. KEI-3 is MG029268.
References
Abreo, N. A. S., Macusi, E. D., Cuenca, G. C., Ranara, C. T. B., Cardona, L., & Arabejo, G. (2015). Nutrient enrichment, sedimentation, heavy metals and plastic pollution in the marine environment and its implications on Philippine marine biodiversity: A Review. IAMURE International Journal of Ecology and Conservation, 15(1), 111-167.
Andersson, M. G. I., Berga, M., Lindström, E. S., & Langenheder, S. (2014). The spatial structure of bacterial communities is influenced by historical environmental conditions. Ecology, 95(5), 1134-1140.
Arora, P. K., Dhar, K., Garc, V., #xed, a, R. A., & Sharma, A. (2015). Biotransformation of Indole to 3-Methylindole by Lysinibacillus xylanilyticus Strain MA. Journal of Chemistry, 2015.
Baum, G., Januar, H. I., Ferse, S. C. A., & Kunzmann, A. (2015). Local and Regional Impacts of Pollution on Coral Reefs along the Thousand Islands North of the Megacity Jakarta, Indonesia. PLOS ONE, 10(9), e0138271.
Bomble, Y. J., Lin, C.-Y., Amore, A., Wei, H., Holwerda, E. K., Ciesielski, P. N., . . . Himmel, M. E. (2017). Lignocellulose deconstruction in the biosphere. Current Opinion in Chemical Biology, 41(Supplement C), 61-70.
Brückner, R., & Titgemeyer, F. (2002). Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS microbiology letters, 209(2), 141-148.
Campbell, B. J., & Kirchman, D. L. (2013). Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. The ISME Journal, 7(1), 210-220.
Chun, J., Lee, J.-H., Jung, Y., Kim, M., Kim, S., Kim, B. K., & Lim, Y.-W. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. International journal of systematic and evolutionary microbiology, 57(10), 2259-2261.
Cleveland, C. C., & Liptzin, D. (2007). C:N:P stoichiometry in soil: is there a “Redfield ratio” for the microbial biomass? Biogeochemistry, 85(3), 235-252. doi:10.1007/s10533-007-9132-0
Coker, J. A. (2016). Extremophiles and biotechnology: current uses and prospects. F1000Research, 5, F1000 Faculty Rev-1396. doi:10.12688/f1000research.7432.1
Cook, G. M., Janssen, P. H., & Morgan, H. W. (1993). Simultaneous uptake and utilisation of glucose and xylose by Clostridium thermohydrosulfuricum. FEMS microbiology letters, 109(1), 55-61.
Esmaeili, A., Pourbabaee, A. A., Alikhani, H. A., Shabani, F., & Esmaeili, E. (2013). Biodegradation of Low-Density Polyethylene (LDPE) by Mixed Culture of Lysinibacillus xylanilyticus and Aspergillus niger in Soil. PLOS ONE, 8(9), e71720.
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., & Olsen, G. J. (2008). Critical Evaluation of Two Primers Commonly Used for Amplification of Bacterial 16S rRNA Genes. Applied and Environmental Microbiology, 74(8), 2461-2470.
Görke, B., & Stülke, J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nature Reviews Microbiology, 6, 613.
Irfan, M., Mushtaq, Q., Tabssum, F., Shakir, H. A., & Qazi, J. I. (2017). Carboxymethyl cellulase production optimization from newly isolated thermophilic Bacillus subtilis K-18 for saccharification using response surface methodology. AMB Express, 7(1), 29.
Juturu, V., & Wu, J. C. (2014). Microbial cellulases: Engineering, production and applications. Renewable and Sustainable Energy Reviews, 33(Supplement C), 188-203.
Karakasidou, K., Nikolouli, K., Amoutzias, G. D., Pournou, A., Manassis, C., Tsiamis, G., & Mossialos, D. (2018). Microbial diversity in biodeteriorated Greek historical documents dating back to the 19th and 20th century: A case study. MicrobiologyOpen, e00596.
Khianngam, S., Pootaeng-on, Y., Techakriengkrai, T., & Tanasupawat, S. (2014). Screening and identification of cellulase producing bacteria isolated from oil palm meal. Journal of Applied Pharmaceutical Science, 4(4), 90.
Kim, J.-H., Shoemaker, S. P., & Mills, D. A. (2009). Relaxed control of sugar utilization in Lactobacillus brevis. Microbiology, 155(4), 1351-1359.
Kuhad, R. C., Deswal, D., Sharma, S., Bhattacharya, A., Jain, K. K., Kaur, A., . . . Karp, M. (2016). Revisiting cellulase production and redefining current strategies based on major challenges. Renewable and Sustainable Energy Reviews, 55, 249-272.
Kuhad, R. C., Gupta, R., & Singh, A. (2011). Microbial Cellulases and Their Industrial Applications. Enzyme Research, 2011, 10.
Kumar, S., Sharma, N., & Pathania, S. (2017). Cost effective production of cellulase using wheat bran from Bacillus subtilis BM1 and encoding endo-beta-1, 4-glucanase producing gene. Molecular biology and evolution, Res. Environ. Life Sci.10(6) 507-512
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870-1874.
L Bergquist, P., W Morgan, H., & Saul, D. (2014). Selected enzymes from extreme thermophiles with applications in biotechnology. Current Biotechnology, 3(1), 45-59.
Lallias, D., Hiddink, J. G., Fonseca, V. G., Gaspar, J. M., Sung, W., Neill, S. P., . . . Creer, S. (2015). Environmental metabarcoding reveals heterogeneous drivers of microbial eukaryote diversity in contrasting estuarine ecosystems. The ISME Journal, 9(5), 1208-1221.
Lee, C. S., Jung, Y.-T., Park, S., Oh, T.-K., & Yoon, J.-H. (2010). Lysinibacillus xylanilyticus sp. nov., a xylan-degrading bacterium isolated from forest humus. International journal of systematic and evolutionary microbiology, 60(2), 281-286.
Lin, C. Y., & Lay, C. H. (2004). Carbon/nitrogen-ratio effect on fermentative hydrogen production by mixed microflora. International Journal of Hydrogen Energy, 29(1), 41-45.
Littlechild, J. A. (2015). Enzymes from Extreme Environments and Their Industrial Applications. Frontiers in Bioengineering and Biotechnology, 3, 161.
Liu, S., Skinner-Nemec, K. A., & Leathers, T. D. (2008). Lactobacillus buchneri strain NRRL B-30929 converts a concentrated mixture of xylose and glucose into ethanol and other products. Journal of industrial microbiology & biotechnology, 35(2), 75-81.
Meddeb-Mouelhi, F., Moisan, J. K., & Beauregard, M. (2014). A comparison of plate assay methods for detecting extracellular cellulase and xylanase activity. Enzyme and Microbial Technology, 66(Supplement C), 16-19.
Moyle, P. B., Lund, J. R., Bennett, W. A., & Fleenor, W. E. (2010). Habitat variability and complexity in the upper San Francisco Estuary. San Francisco Estuary and Watershed Science,
8(3).
Poli, A., Finore, I., Romano, I., Gioiello, A., Lama, L., & Nicolaus, B. (2017). Microbial Diversity in Extreme Marine Habitats and Their Biomolecules. Microorganisms, 5(2).
Reed, H. E., & Martiny, J. B. H. (2013). Microbial composition affects the functioning of estuarine sediments. The ISME Journal, 7(4), 868-879.
Rojas-Pinzón, P. A., & Dussán, J. (2017). Efficacy of the vegetative cells of Lysinibacillus sphaericus for biological control of insecticide-resistant Aedes aegypti. Parasites & Vectors, 10(1), 231.
Sindhu, R., Binod, P., & Pandey, A. (2016). Biological pretreatment of lignocellulosic biomass–An overview. Bioresource technology, 199, 76-82.
Stierle, A. A., & Stierle, D. B. (2014). Bioactive Secondary Metabolites from Acid Mine Waste Extremophiles. Natural product communications, 9(7), 1037-1044.
Subudhi, H., & Patra, H. K. (2013). Mangrove forests of river estuaries of Odisha, India. International Journal of Biodiversity and Conservation, 5(8), 446-454.
Sujana, K. A., Saravanan, R., & Pandey, A. D. (2015). Distribution of Aquatic Macrophytes in Balasore District, Odisha. In M. Rawat, S. Dookia, & C. Sivaperuman (Eds.), Aquatic Ecosystem: Biodiversity, Ecology and Conservation (pp. 1-12). New Delhi: Springer India.
Sun, Z., Li, G., Wang, C., Jing, Y., Zhu, Y., Zhang, S., & Liu, Y. (2014). Community dynamics of prokaryotic and eukaryotic microbes in an estuary reservoir. Scientific Reports, 4, 6966.
Wheeler, P. A., & Kirchman, D. L. (1986). Utilization of inorganic and organic nitrogen by bacteria in marine systems. Limnology and Oceanography, 31(5), 998-1009.
Yin, J., Chen, J.-C., Wu, Q., & Chen, G.-Q. (2015). Halophiles, coming stars for industrial biotechnology. Biotechnology advances, 33(7), 1433-1442.
Zhang, W., Bougouffa, S., Wang, Y., Lee, O. O., Yang, J., Chan, C., . . . Qian, P.-Y. (2014). Toward Understanding the Dynamics of Microbial Communities in an Estuarine System. PLOS ONE, 9(4), e94449.