Department of Biotechnology, SAGE University, Indore – 452020, India
Corresponding author email: sidrahh75@gmail.com
Article Publishing History
Received: 15/07/2025
Accepted After Revision: 28/09/2025
The OsRFP45 identified in Oryza sativa has been identified as a negative salt-stress regulator. Its functions have been studied previously by overexpression and knock-out lines, but it has not been experimentally validated in structural form. This study provides a homology-based three-dimensional structural prediction of OsRFP45, an E3 ubiquitin ligase discovered in Oryza sativa, based on a matching template using SWISS-MODEL.Based on the existing literature, the locus Id of the OsRFP45 was identified. The locus Id was searched on NCBI to obtain the FASTA file of the identified protein. NCBI BLAST was conducted and it was analysed that the gene responsible for the production of this protein is highly conserved among various species. The homology modelling of the RING domain of OsRFP45 was performed using a template which had the highest GMQE (Global Model Quality Estimate). The accuracy of the predicted structural model was validated by running the bioinformatics tool – ProCheck and obtaining the Ramachandran Plot and related parameters. According to sequence alignment, OsRFP45’s conserved RING domain is highly comparable to proteins from the RING domain found in other plant species.The OsRFP45 RING domain’s conserved structural characteristics align with classical C3HC4-type E3 ubiquitin ligase. The three-dimensional structure shows a compact RING-domain stabilised by conserved cysteine and histidine residues, indicating a zinc-coordinated fold required for E3 ligase function. Zero percent of the disallowed region, 85.5% of the core and 14.5% of the allowed region are concluded by the Ramachandran plot. The predicted structure is hence, confirmed as correct.
BLAST, Conserved-RING OsRFP45, Homology Modelling,
Hypothetical protein, Multiple sequence alignment.
Hashmi S, Pillai L. Homology Modelling and Evaluation of a Conserved Domain in Oryza sativa RING Finger Protein 45, a Putative E3 Ubiquitin Ligase. Biosc.Biotech.Res.Comm. 2025;18(3).
Hashmi S, Pillai L. Homology Modelling and Evaluation of a Conserved Domain in Oryza sativa RING Finger Protein 45, a Putative E3 Ubiquitin Ligase. Biosc.Biotech.Res.Comm. 2024;18(3). Available from: <a href=”https://shorturl.at/uVu2v“>https://shorturl.at/uVu2v</a>
INTRODUCTION
Over half of the world’s population depends on rice (Oryza sativa L.), which is also the most significant crop in Asian nations. Africa has also seen a significant increase in rice consumption recently (Seck et al., 2012). The major abiotic stress which is responsible for poor yield and quality of grain is salinity. The accumulation of salt in soil causes stress conditions, due to this 10% of global cultivable land is not available for growth.( Saleem et al., 2025). The current estimate of saline land globally is 800 million hectares, with the amount increasing by 1% to 2% annually (Munns and Tester, 2008).
Crop growth and development are greatly impacted by soil salinization, which also depletes soil nutrients and breaks down the structure of soil particles (Kordrostami, Rabiei and Kumleh, 2017). Rising sea levels brought on by global warming exacerbate salinity stress, which lowers soil fertility and hinders crop development in coastal areas (Shrivastava and Kumar, 2014). Rice grain yield encounters a sharp decline due to various forms of unavoidable biotic and abiotic stresses (Dixit et al., 2020 ; Khush, 2005). Previous studies have concluded that there is a nearly 29.29% decline in rice yield in soils with high salt accumulation in contrast to non-saline soils (Oelviani et al., 2024).
Since it is a glycophyte by nature, too much salt adversely impacts its growth. Studying how rice reacts specifically to ion build-up above the threshold level can help identify the main causes of stunted growth and low rice yield, as well as the potential for future mitigation of the same (Ghosh and Ali, 2016). A RING-v-type E3 ubiquitin ligase has recently been identified, called OsRFP45 (Oryza sativa RING finger Protein 45). Present on chromosome 6, it has been concluded that it plays a major role in the negative regulation of salt tolerance in Oryza sativa. (Choi, Kim and Jang, 2025).
Plants have established post-translational mechanisms for regulation to combat the various stresses encountered, and the ubiquitin–proteasome system (UPS) is an essential component of these systems. Attaching ubiquitin molecules to target proteins and designating them for 26S proteasome destruction is known as ubiquitination. E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase) are the three enzymes that control this process. Rice’s ability to withstand salt stress has been connected to a number of E3 ubiquitin ligases with conserved RING domains. Regulation of plant stress responses and determining specificity for the substrate depend on E3 ubiquitin ligases. Based on their structure, E3 ubiquitin ligases are further grouped into single and multi-subunit divisions. U-box, homology to the E6AP C terminus (HECT), and Really Interesting New Gene (RING) are the different groups of single-subunit E3 ubiquitin ligases ( Deshaies and Joazeiro, 2009, Lee and Kim, 2011, Choi, Kim and Jang, 2025).
The plant stress hormone widely associated with response to environmental stimuli is Abscisic Acid (ABA) (Yamaguchi-Shinozaki and Shinozaki, 2006; Zhu, 2002 ). It has been observed that cellular ABA levels rapidly increase under adverse circumstances, which include a variety of outcomes, comprising – restriction of growth, closing of stomatal pores, sugar accumulation and generation of proline in significant amounts. Abscisic acid synthesis and the stress response it mediates are regulated by distinct E3 Ligases (Ko, Yang and Han, 2006). RING E3 Ubiquitin ligases are present abundantly in eukaryotic organisms. Rice and Arapidopsis consist of 378 and 499 genes, respectively in contrast to Saccharomyces cerevisiae which involves, 47 genes that encode for RING-type E3. (Kraft et al., 2005; Mazzucotelli et al., 2006). OsRFP45, a v-type RING E3 ligase, was identified as a prospective regulator of salt stress responses because of its transcriptional modification as an outcome of abiotic stresses such as drought and increased salt accumulation(Choi, Kim and Jang, 2025).
Previous studies examined into the way OsRFP45 controls ROS build-up, Na+ and K+ homeostasis, and its control of gene expression associated with stress under salt-induced stress (Choi, Kim and Jang, 2025). However, OsRFP45’s structural characterisation is still limited because there isn’t a 3D structure that has been experimentally determined yet. In order to shed light on the conserved C3HC4-type RING finger domain of OsRFP45 and its possible function in modulating protein–protein interactions and E3 ligase activity, the present study provides an in silico analysis of its structure. Homology modelling was performed to build a predictive 3D structure of OsRFP45’s conserved RING finger domain, with the goal of acquiring a better understanding of its structural properties and functional properties.
MATERIAL AND METHODS
Numerous proteins’ sequences can now be ascertained by efficiently recognising, isolating, and sequencing genes owing to the development of molecular biology tools. The protein structure is usually determined by X-ray crystallography, followed by Nuclear Magnetic Resonance. However, due to the increase in research and development of genomics there is an inevitable gap between the scientifically confirmed 3D model structures and the protein sequences found. In Silico techniques offer a solution to this gap by providing more rapid, cost-effective, and practical outcomes. Using previously determined structures of related proteins as templates, homology modelling is a computational technique that predicts the three-dimensional arrangement of proteins based on their amino acid sequences. It is regarded as the most accurate of computational structure prediction techniques (Bishop et al., 2008; Muhammed and Aki‐Yalcin, 2018).
Sequence Retrieval : The OsRFP45 was identified as LOC_Os06g46366 (Choi, Kim and Jang, 2025). This was identified using the MSU Rice Genome Annotation Project, for cross-referencing the RAP-DB ID was obtained which was used to obtain the protein sequence from NCBI database. This retrieved the complete protein sequence, to get the exact RING sequence, InterProScan was used which annotated the exact sequence of the conserved RING region. The BLASTp alignment graphical summary confirmed that the RING finger domain, specifically the C4HC3-type protein was found out to be present at 241-289. This domain is involved in E3 ubiquitin ligase activity, typically mediating protein degradation. To accurately perform the homology modelling the range of (protein sequence) 231-300 amino acids was selected and saved as FASTA file.
The main phases involved in modelling a three-dimensional protein structure from its sequence: choosing a template to use, aligning the target and template, developing structural models, and assessing the models. SWISS-MODEL was chosen for homology modelling due to its reliability and continuous advancement throughout automated comparison modelling. The program is especially well-suited for producing precise 3D structures even in the absence of experimentally determined models since it makes use of large template libraries and advanced quality estimation metrics. In order to improve model accuracy and interpretability, its most current improvements include better protein assembly modelling capabilities and more biologically relevant template selection techniques (Waterhouse et al., 2018).
The FASTA file was first uploaded to SWISS-MODEL to find the matching template. The match having the highest GMQE (Global Model Quality Estimate) was AlphaFold DB model A0A0D2QDE7.1. A RING-CH-type domain-containing protein which showed 0.89 GMQE. It resulted in 92.86% Seq. Identity. The Oligo-State of the template was monomer. This template was hence, chosen to accurately model the RING-domain of OsRFP45 protein.
Figure 1: Homology model of the RING finger domain of OsRFP45 (XP_015644308.1).
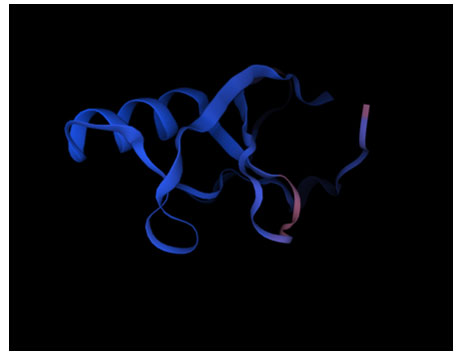
Model Assessment :The quality of the initial alignment and resulting sequence identity will have a significant impact on the model’s final quality (Martí-Renom et al., 2000). There are various methods for determining how accurate a model is, including geometrical inspections, empirical energy functions, and statistical grading. A protein structural model’s utility is determined by its degree of precision. It should be attainable to produce a high-quality model structure, if a known protein matches more than 60% of its identity with the sequence being investigated (Chothia and Lesk, 1986). The protein template used (gene : A0A0D2QDE7_GOSRA, organism: Gossypium raimnodii (Peruvian cotton) ( Gossypium klotzschianum subspraimondii) resulted in 92.86% identity, hence the homology model showed an accurate result.
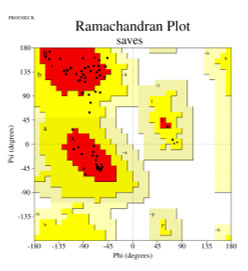
The modelled OsRFP45 domain was ran through several validation procedures through the SAVES server. ERRAT, which analyses non-bonded atom-to-atom interactions using a 9-residue sliding window, was used to evaluate the model’s quality (Colovos and Yeates, 1993). It generates an overall quality factor based on comparisons to high-resolution structures; scores above 90% are suggestive of dependable models. It had an ERRAT quality score of 97.727%, which suggests that the predicted model had a high stereochemical quality. The ERRAT score confirms the accuracy of the model by indicating that there are no major areas of structural defects in the backbone and side chain environment. PROCHECK is a renowned bioinformatics tool for assessment of the reliability of stereochemical characteristics of three-dimensional structures of protein. One of its primary outputs is the Ramachandran plot, which illustrates the spatial distribution of the backbone dihedral angles—phi (ϕ) and psi (ψ)—of all the amino acid residues in the protein structure. These angles determine the configuration of the polypeptide chain. The representation separates residues into most favoured, additionally allowed, generously allowed, and disallowed regions according to known steric constraints.
A high proportion of residues in preferred locations indicates a reliable and accurately folded protein model. The Ramachandran plot is a key validation step used to evaluate the modelled structure’s overall reliability and stereochemical stability(Laskowski et al., 1993; Ramachandran, Ramakrishnan and Sasisekharan, 1963). The PSIPRED bioinformatics tool (Jones, 1999), which is well-known for its excellent accuracy in predicting α-helices, β-strands, and coil regions from the primary amino acid sequence, was used to further validate the secondary structure prediction of the modelled protein.
A further level of validation was provided by the PSIPRED output (Figure 3), which allowed a comparison between the secondary structural elements predicted and those seen in the homology model. The created three-dimensional model’s dependability and appropriateness for additional investigations like docking or functional annotation are further enhanced by a high degree of agreement between the expected and modelled secondary structures.
RESULTS AND DISCUSSION
Figure 2: ERRAT Overall Quality Factor – 97.727 %
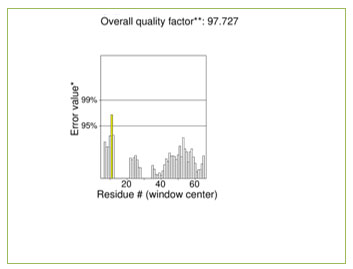
Figure 3: PSIPRED Secondary structure
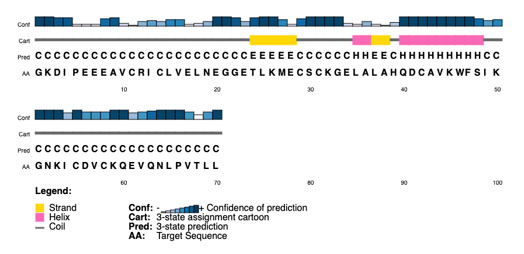
Figure 4: Ramachandran plot
E3 ubiquitin ligases are characterized by a compact RING-domain in the expected three-dimensional structure, which is held in place by conserved cysteine and histidine residues that are responsible for coordination of zinc ions (Figure 1). The conserved RING domain’s correct fold and functional conformation depend on this zinc-chelating motif.
Using the PROCHECK tool, the model’s stereochemical quality was evaluated. Zero percent of the disallowed region, 85.5% of the core and 14.5% of the allowed region are shown by the Ramachandran plot. The predicted structure is hence, confirmed. According to this distribution, the model’s stereochemical validity is supported and the anticipated structure complies well with established dihedral angle values (Figure 1).
Additionally, PSIPRED’s projected secondary structure elements closely matched the modelled structure’s observed elements, demonstrating a strong alignment of loops, β-strands, and α-helices. The structural accuracy of the homology model is further supported by this alignment. When combined, the PROCHECK and PSIPRED validation results verify that the modelled structure is accurate and suitable for additional functional or docking research.
CONCLUSION
In order to examine the spatial organization of conserved cysteine and histidine residues, the homology model of the putative RING finger protein OsRFP45 from Oryza sativa was built using SWISS-MODEL. The lack of experimentally determined structural data for OsRFP45 led to the pursuit of homology modelling. The study concentrated on the RING domain because the full-length protein’s template alignment was not optimal. This region binds two zinc atoms necessary for structural integrity and ligase function. It is well-known for its chelating residues, which are mainly cysteine and histidine. Validation metrics showed a structurally sound model appropriate for additional docking and functional investigations, while the predicted 3D model showed a well-structured RING domain with conserved zinc-coordinating residues.
The conserved RING domain is represented by the 241–289 amino acid region, as validated by the BLASTp graphical summary. Furthermore, a secondary structure with short α-helices and β-strands scattered with coil regions was predicted by PSIPRED. These regions most likely represent flexible loops that support domain stability. When taken as a whole, OsRFP45’s conserved structural characteristics provide compelling evidence for its designation as an E3 ubiquitin ligase. This model establishes the foundation for upcoming mutational and in silico docking investigations that may aid in elucidating OsRFP45’s function in rice stress response networks.
ACKNOWLEDGMENTS
The authors are highly thankful to the Department of Biotechnology, SAGE University, Indore, India for providing support in the form of Bioinformatics infrastructure facility to carry out this work.
Conflict of Interest Statement: The authors declare that there is no conflict of interest.
Data Availability Statement: Data are available with the corresponding author on a reasonable request.
REFERENCES
Barlow, P.N. B Luisi, A Milner, M Elliott, R Everett (1994) Structure of the C3HC4 domain by 1H-Nuclear magnetic resonance spectroscopy. Journal of Molecular Biology, 237(2), pp. 201–211. https://doi.org/10.1006/jmbi.1994.1222
Bishop, A., Tastan, A.O., de Beer, T.A.P. and Joubert, F. (2008) ‘Protein homology modelling and its use in South Africa’, South African Journal of Science, 104(1–2), pp. 2–6. Available at: http://www.scielo.org.za/scielo.php?script=sci_arttext&pid=S0038-23532008000100001
Borden, K.L. and Freemont, P.S. (1996) ‘The RING finger domain: a recent example of a sequence—structure family,’ Current Opinion in Structural Biology, 6(3), pp. 395–401. https://doi.org/10.1016/s0959-440x(96)80060-1.
Choi, M.S., Kim, J.H. and Jang, C.S. (2025) ‘Rice RING E3 Ligase OSRFP45 negatively regulates salt tolerance by modulating NA+/K+ transporter genes,’ Physiologia Plantarum, 177(3). https://doi.org/10.1111/ppl.70327.
Chothia, C. and Lesk, A.M. (1986) ‘The relation between the divergence of sequence and structure in proteins.,’ The EMBO Journal, 5(4), pp. 823–826. https://doi.org/10.1002/j.1460-2075.1986.tb04288.x.
Colovos, C. and Yeates, T.O. (1993) ‘Verification of protein structures: Patterns of nonbonded atomic interactions,’ Protein Science, 2(9), pp. 1511–1519. https://doi.org/10.1002/pro.5560020916.
Deshaies, R.J. and Joazeiro, C.A.P. (2009) ‘RING Domain E3 Ubiquitin ligases,’ Annual Review of Biochemistry, 78(1), pp. 399–434. https://doi.org/10.1146/annurev.biochem.78.101807.093809.
Dixit, S., Singh, U.M., Singh, A.K., Alam, S., Venkateshwarlu, C., Nachimuthu, V.V. et al., 2020. Marker assisted forward breeding to combine multiple biotic-abiotic stress resistance/tolerance in rice. Rice, 13, pp.1–15. https://doi.org/10.1186/s12284-020-00391-7
Ghosh, B. and Ali, N., MD (2016) ‘Response of Rice under Salinity Stress: A Review Update,’ Rice Research Open Access, 4(2).
https://doi.org/10.4172/2375-4338.1000167.
Hershko, A. and Ciechanover, A. (1998) ‘THE UBIQUITIN SYSTEM,’ Annual Review of Biochemistry, 67(1), pp. 425–479. https://doi.org/10.1146/annurev.biochem.67.1.425.
Khush, G.S. (2005) ‘What it will take to Feed 5.0 Billion Rice consumers in 2030,’ Plant Molecular Biology, 59(1), pp. 1–6. https://doi.org/10.1007/s11103-005-2159-5.
Ko, J., Yang, S.H. and Han, K. (2006) ‘Upregulation of an Arabidopsis RING‐H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis,’ The Plant Journal, 47(3), pp. 343–355. https://doi.org/10.1111/j.1365-313x.2006.02782.x.
Kordrostami, M., Rabiei, B. and Kumleh, H.H. (2017) ‘Biochemical, physiological and molecular evaluation of rice cultivars differing in salt tolerance at the seedling stage,’ Physiology and Molecular Biology of Plants, 23(3), pp. 529–544. https://doi.org/10.1007/s12298-017-0440-0.
Kraft, E., Stone, S.L., Ma, L., Su, N., Gao, Y., Lau, O. et al. (2005) ‘Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis‘, Plant Physiology, 139(4), pp. 1597–1611. https://doi.org/10.1104/pp.105.067983.
Laskowski, R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) ‘PROCHECK: a program to check the stereochemical quality of protein structures’, Journal of Applied Crystallography, 26(2), pp. 283–291. https://doi.org/10.1107/S0021889892009944.
Lee, J.-H. and Kim, W.T. (2011) ‘Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis,’ Molecules and Cells, 31(3), pp. 201–208. https://doi.org/10.1007/s10059-011-0031-9.
Martí-Renom, M.A., Stuart, A.C., Fiser, A., Sánchez, R., Melo, F. and Sali, A. (2000) ‘Comparative protein structure modelling of genes and genomes’, Annual Review of Biophysics and Biomolecular Structure, 29, pp. 291–325. https://doi.org/10.1146/annurev.biophys.29.1.291
Mazzucotelli, S.B., Maronem, D., De Leonardis, A.M., Guerra, D., Di Fonzo, N., Cattivelli, L. et al. (2006) ‘The E3 ubiquitin ligase gene family in plants: regulation by degradation’, Current Genomics, 7(8), pp. 509–522. https://doi.org/10.2174/138920206779315728.
Muhammed, M.T. and Aki‐Yalcin, E. (2018) ‘Homology modeling in drug discovery: Overview, current applications, and future perspectives,’ Chemical Biology & Drug Design, 93(1), pp. 12–20. https://doi.org/10.1111/cbdd.13388.
Munns, R. and Tester, M. (2008) ‘Mechanisms of salinity tolerance,’ Annual Review of Plant Biology, 59(1), pp. 651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911.
Oelviani, R., Adiyoga, W., Suhendrata, T., Bakti, I.G.M.Y., Sutanto, H.A., Fahmi, D.A., Chanifah, C., Jatuningtyas, R.K., Samijan, S., Malik, A. and Sahara, D., 2024. Effects of soil salinity on rice production and technical efficiency: Evidence from the northern coastal region of Central Java, Indonesia. Case Studies in Chemical and Environmental Engineering, 10, p.101010. https://doi.org/10.1016/j.cscee.2024.101010
Pillai, L. and Pardasani, K.R. (2012) ‘Modeling Calcium Dependent Protein Kinase Isoform 1 from Cicer arietinum (chick pea),’ Journal of Computer Science & Systems Biology, 05(01). https://doi.org/10.4172/jcsb.1000084.
Saleem, M.A., Khan, A., Tu, J., et al. (2025) ‘Salinity stress in rice: Multilayered approaches for sustainable tolerance, International Journal of Molecular Sciences, 26(13), p. 6025. https://doi.org/10.3390/ijms26136025.
Seck, P.A., Diagne, A., Mohanty, S. and Wopereis, M.C.S., 2012. Crops that feed the world 7: Rice. Food Security, 4(1), pp.7–24. https://doi.org/10.1007/s12571-012-0168-1
Shrivastava, P. and Kumar, R. (2014) ‘Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation,’ Saudi Journal of Biological Sciences, 22(2), pp. 123–131. https://doi.org/10.1016/j.sjbs.2014.12.001.
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F.T., de Beer, T.A.P., Rempfer, C., Bordoli, L., Lepore, R. and Schwede, T., 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), pp.W296–W303. https://doi.org/10.1093/nar/gky427
Yamaguchi-Shinozaki, K. and Shinozaki, K. (2006) ‘Transcriptional Regulatory Networks In Cellular Responses And Tolerance To Dehydration And Cold Stresses,’ Annual Review of Plant Biology, 57(1), pp. 781–803. https://doi.org/10.1146/annurev.arplant.57.032905.105444.
Zhu, J.-K. (2002) ‘Salt And Drought Stress Signal Transduction In Plants,’ Annual Review of Plant Biology, 53(1), pp. 247–273. https://doi.org/10.1146/annurev.arplant.53.091401.143329.


