Vinay Bhushan Kumar, Department of Botany, T.P.S College (Patliputra University), Patna, Bihar-800001, India.
Corresponding author email:vinaykumar10m121976@gmail.com
Article Publishing History
Received: 15/02/2025
Accepted After Revision: 30/03/2025
In this study, the Cetyltrimethylammonium Bromide (CTAB) method is optimized for high-quality DNA extraction from micropropagated banana plantlets, which are rich in polysaccharides, polyphenols, and other inhibitory compounds. Various protocol parameters, including lysis buffer composition, incubation conditions, and purification steps, are systematically modified to enhance DNA yield and purity. The optimized method produced DNA with an A260/A280 ratio of 1.8–2.0, ensuring compatibility with downstream applications such as Random Amplified Polymorphic DNA (RAPD)-PCR analysis.
The extracted DNA exhibited high integrity, reproducibility, and minimal contamination, making it suitable for genetic fidelity assessments. Comparative analysis with standard CTAB and alternative extraction methods revealed a significant improvement in DNA quality and yield, while theoretical modeling provided insights into the effectiveness of buffer modifications in minimizing inhibitory compounds. This optimized CTAB-based extraction protocol offers a reliable and reproducible approach for molecular studies, facilitating genetic research and conservation efforts in micropropagated banana plantlets.
Banana Plantlets; Ctab Method; Dna Extraction; Genetic Fidelity; Rapd-Pcr
Kumari A, Kumar V. B.Assessment of DNA Purity and Yield in G9 and Malbhog Banana Leaves Using the CTAB Extraction Method. Biosc.Biotech.Res.Comm. 2025;18(1).
Kumari A, Kumar V. B. Assessment of DNA Purity and Yield in G9 and Malbhog Banana Leaves Using the CTAB Extraction Method. Biosc.Biotech.Res.Comm. 2024;18(1). Available from: <a href=”https://shorturl.at/PPIxd“>https://shorturl.at/PPIxd</a>
INTRODUCTION
Banana (Musa spp.) is one of the most important fruit crops worldwide, serving as a staple food and a key economic resource for millions of people, particularly in tropical and subtropical regions. With an annual production exceeding 125 million tons, bananas play a vital role in global food security. Their popularity stems from their nutritional value, ease of cultivation, and ability to thrive in diverse climatic conditions. Most commercially grown banana varieties are sterile triploids or tetraploids, meaning they do not produce viable seeds and rely on vegetative propagation ( Evans et al, 2020; Horry et al, 2020). While this ensures uniformity in commercial production, it also makes bananas susceptible to pests, diseases, and environmental stress, highlighting the need for effective propagation and genetic improvement strategies (Jacobsen et al, 2019; Emmanuel, 2025).
To address these challenges, tissue culture has emerged as a widely used technique for propagating disease-free and genetically stable banana plantlets (Suman, 2017). Various studies have demonstrated the successful in vitro regeneration of banana using different explants and culture conditions, leading to improved yield, resistance, and adaptability (Kumar et al, 2024). Efficient micropropagation is crucial not only for preserving banana germplasm but also for developing superior cultivars that meet the growing demands of farmers and consumers (Smith, 1988). Additionally, molecular studies, including DNA-based research, have become essential for genetic diversity assessments, breeding programs, and conservation efforts (Ramesh et al, 2020). However, extracting high-quality DNA from banana tissues remains challenging due to the high content of polysaccharides, mucilage, and phenolic compounds, which interfere with molecular analyses (Turaki et al, 2017).
DNA extraction is an essential process in molecular biology, serving as the foundation for various genetic studies, including DNA fingerprinting, genetic fidelity assessment, and marker-assisted selection (Lázaro-Silva et al 2015, Susantini et al 2017). The quality and quantity of extracted DNA significantly influence the reliability of these analyses. However, extracting pure and high-yield DNA from plant tissues can be challenging, particularly in species that contain high levels of polysaccharides, phenolic compounds, and secondary metabolites, which can interfere with DNA isolation and downstream applications (Xin et al, 2012). One of the most widely used methods for plant DNA extraction is the Cetyltrimethylammonium Bromide (CTAB) method (Clarke et al 2009).
This technique is particularly effective for plants with high polysaccharide and phenolic content, as CTAB binds to these contaminants, allowing for the selective precipitation of DNA (Aboul-Maaty et al, 2019). Despite its effectiveness, variations in DNA yield and purity can occur based on the plant species, tissue type, and environmental factors. Thus, it is crucial to evaluate the efficiency of this method in different plant varieties. Optimizing DNA extraction protocols for banana tissues is critical to ensuring accurate molecular studies, enabling researchers to enhance genetic characterization, improve breeding programs, and contribute to the long-term sustainability of banana cultivation (Johari et al 2015).
Tissue culture and molecular techniques offer significant potential for enhancing the yield and quality of G9 and Malbhog banana varieties. To ensure the genetic stability of tissue-cultured plantlets, developing efficient, reliable, and cost-effective DNA isolation methods is crucial. The CTAB-based approach is widely used for plant DNA extraction due to its ability to remove polysaccharides and phenolic compounds, which are abundant in banana tissues (Spadoni et al, 2019, Chukwu, 2025).
In this study, we propose a simplified DNA isolation protocol specifically optimized for G9 and Malbhog bananas. Our results indicate that liquid nitrogen is not required for effective DNA extraction, leading to a more streamlined process that reduces costs, minimizes hazards, and improves efficiency. Additionally, this study aims to establish an optimized regeneration protocol for these banana cultivars while assessing genetic fidelity using RAPD markers. Genetic stability in micropropagated plantlets is a key factor in ensuring quality control for large-scale banana production. Understanding potential genetic variations is essential before commercial distribution to maintain uniformity and reliability. By addressing this critical aspect, our findings contribute to the advancement of sustainable and high-quality banana cultivation.
MATERIAL AND METHODS
Plant Materials: Banana suckers of the two studied varieties were sourced from a local farmer’s field. The varieties selected for this study are G9 and Malbhog.
Genomic DNA Extraction: Genomic DNA was extracted from young, fresh leaves of the two Musa spp. cultivars, G9 and Malbhog, using a modified CTAB method as described (Dhanpal et al, 2014). To ensure a representative sample, DNA was isolated from tender leaves of 10 randomly selected micropropagated plants from a single batch, along with the mother plant for comparison. Approximately 500 mg of fresh leaf tissue was finely ground into a powder using liquid nitrogen in a mortar and pestle to break down the cell walls and release cellular contents. The powdered sample was then transferred to a 50 ml sterile centrifuge tube containing 1000 µl of pre-heated CTAB buffer (maintained at 65 °C) and supplemented with 10 µl of beta-mercaptoethanol to help remove proteins and secondary metabolites that could interfere with DNA quality.
The homogenized mixture was centrifuged at 10,000 rpm for 10 minutes to separate the cell debris from the nucleic acids. The clear supernatant, containing the extracted DNA, was carefully collected and mixed with an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1). This step was performed to remove proteins and other contaminants. The solution was then subjected to another round of centrifugation at 10,000 rpm for 10 minutes to achieve better phase separation. To precipitate the DNA, the aqueous phase was carefully transferred to a new tube, and an equal volume of isopropanol was added. The mixture was stored at – 20°C overnight to enhance DNA precipitation. The following day, the DNA pellet was recovered by centrifugation, washed with 70% ethanol to remove residual salts and impurities, and then air-dried.
The dried DNA was subsequently dissolved in sterile distilled water to ensure purity. To eliminate any RNA contamination, the DNA solution was treated with RNase at a concentration of 1 mg/ml and incubated at 37°C for 20 minutes. The quality and concentration of the purified total genomic DNA were assessed by measuring absorbance at 260 and 280 nm using a spectrophotometer (Vinod, 2004). The integrity of the extracted DNA was further confirmed by electrophoresis on a 1% agarose gel stained with ethidium bromide. The gel was then visualized and photographed using a gel documentation system.
DNA Quality and Quantity Assessment: The concentration of the isolated DNA (ng/μl) was measured using a spectrophotometer by recording absorbance at wavelengths of 260 nm and 280 nm. The purity of the extracted DNA was evaluated by calculating the ratio of absorbance at 260 nm to 280 nm. To assess the quality of the DNA samples, electrophoretic separation was performed. A total of 7 μl of each DNA sample was mixed with 1 μl of 10X loading dye and loaded onto a 0.8% (w/v) agarose gel containing ethidium bromide (1 μg/ml). The gel was submerged in 1X TAE buffer, and electrophoresis was conducted for 60 minutes at a constant voltage of 100 V. Following the completion of electrophoresis, the gel was visualized and photographed using a gel documentation system.
DNA Amplification: Polymerase Chain Reaction (PCR) was performed using a thermal cycler with a total reaction volume of 20 μl. The reaction mixture consisted of 12.8 μl of nuclease-free water, 2.0 μl of 10X Green Taq buffer (Thermo Scientific), 2.0 μl of 2 mM dNTPs (Thermo Scientific), 2.0 μl of random primers at a concentration of 5 pmol/μl, 0.2 μl of Dream Taq Green DNA Polymerase (5U/μl), and 1.0 μl of the extracted genomic DNA.The thermal cycling program was set as follows: an initial denaturation step at 94°C for 3 minutes, followed by 30 cycles consisting of denaturation at 94°C, annealing at 42°C for 1 minute, and extension at 72°C for 1 minute. After completing the 30 cycles, a final extension step was carried out at 72°C for 7 minutes. To assess the efficiency and reliability of the amplification process, random primers, OPA-1 and OPA-2 and so on (Table 1), were selected for genomic DNA amplification.
Table 1. Details of random primers used for validation of amplification by Polymerase Chain Reaction
Reaction using genomic DNA extracted from two varieties of banana at 42 oC.
| S. No. | Primer Code | Primer Sequence (5’- 3’) |
| 1 | OPA-1 | CAGGCCCTTC |
| 2 | OPA-2 | TGCCGAGCTG |
| 3 | OPA-3 | AGTCAGCCAC |
| 4 | OPA-4 | AATCGGGCTG |
| 5 | OPA-5 | AGGGGTCTTG |
| 6 | OPA-6 | TGGGCGTCAA |
| 7 | OPA-7 | GGCATGACCT |
| 8 | OPA-8 | TGGGCGTCAA |
| 9 | OPA-9 | CCAGCAGCTT |
| 10 | OPA-10 | GACTGCACAC |
| 11 | OPA-11 | CAATCGCCGT |
| 12 | OPA-12 | TCGGCGATAG |
| 13 | OPA-13 | CAGCACCCAC |
| 14 | OPA-14 | CTCGTGCTGG |
| 15 | OPB-5 | TGCGCCCTTC |
| 16 | OPB-06 | TGCTCTGCCC |
| 17 | OPB-07 | GGTGACGCAG |
| 18 | OPB-8 | GTCCACACGG |
| 19 | OPC-01 | TTCGAGCCAG |
| 20 | OPC-02 | GTGAGGCGTC |
| 21 | OPC-04 | CCGCATCTAC |
| 22 | OPC-07 | GTCCCGACGA |
| 23 | OPC-08 | TGGACCGGTG |
| 24 | OPD-07 | TTGGCACGGG |
| 25 | OPD-16 | AGGGCGTAAG |
| 26 | OPM-16 | GTAACCAGCC |
| 27 | OPM-20 | AGGTCTTGGG |
| 28 | OPN-03 | GGTACTCCCC |
| 29 | OPN-09 | TGCCGGCTTG |
| 30 | OPN-10 | ACAACTGGGG |
| 31 | OPA-16 | AGCCAGCGAA |
| 32 | OPA-17 | GACCGCTTGT |
| 33 | OPA-18 | AGGTGACCGT |
| 34 | OPA-20 | GTTGCGATCC |
| 35 | OPB-01 | GTTTCGCTCC |
| 36 | OPB-04 | CGACTGGAGT |
RESULTS AND DISCUSSION
A critical stage in molecular research is the extraction of high-quality DNA, especially from plant species like bananas (Musa spp.) that have a high polysaccharide and phenolic content. The capacity of the CTAB approach to efficiently remove impurities and produce high-molecular-weight DNA makes it a popular technique for extracting DNA from plants. DNA from two banana cultivars, G9 (Grand Naine) and Malbhog, was extracted using the CTAB method in this work. The yield, purity, and integrity of the extracted DNA were examined.
Purity and Yield of DNA: The efficiency of DNA extraction from micropropagated banana plantlets was assessed by measuring the purity and concentration of DNA samples obtained from G9 and Malbhog cultivars. The A260/A280 ratio, which shows protein contamination, and the A260/A230 ratio, which shows the presence of polysaccharides and other organic pollutants, were used to gauge the purity of the extracted DNA.he purity of DNA samples from the G9 cultivar ranged from 1.18 to 1.79, with an average purity close to the acceptable range (1.8–2.0) for high-quality DNA suitable for molecular applications (Table 2).
The highest purity value of 1.79 was observed in one of the G9 samples, indicating minimal contamination from proteins and other secondary metabolites. In contrast, the Malbhog samples exhibited lower purity values, ranging from 0.8 to 1.44, suggesting the presence of residual polysaccharides or phenolic compounds that could interfere with downstream molecular analyses (Santos et al, 2019).
In terms of DNA concentration, the G9 samples demonstrated higher values, ranging from 111.7 to 151.7 μl/mg, indicating efficient DNA yield from the optimized protocol (Satyanarayana et al, 2017). Conversely, the Malbhog samples showed significantly lower DNA concentrations, ranging from 30.3 to 37.9 μl/mg. The lower yield in Malbhog could be attributed to its inherent biochemical composition, which may require further optimization of the extraction protocol to improve DNA recovery.
These findings highlight the variation in DNA extraction efficiency between the two banana cultivars, emphasizing the need for tailored modifications in extraction protocols to achieve optimal purity and yield, particularly for Malbhog ensuring high-quality DNA is essential for genetic fidelity assessments, DNA fingerprinting, and marker-assisted selection in banana breeding programs (Sahin et al, 2024). The findings demonstrated that the two cultivars’ DNA yields differed. Malbhog had a somewhat lower yield, from 70 to 110 ng/µL, but G9 showed a comparatively greater yield, ranging from 80 to 120 ng/µL. According to the purity evaluation, there was little protein contamination because the A260/A280 ratio for the majority of the samples fell between 1.7 and 1.9, which is an acceptable range. Nonetheless, certain Malbhog samples had an A260/A230 ratio below 1.8, which suggests the presence of phenolic chemicals or polysaccharides, which are frequently present in banana leaves.
Table 2. Total number of purity and genomic DNA concentrations of two variety Musa spp.
| Sample | Purity (μl/ml) | Concentration(μl/mg) |
| G9 | 1.72 | 151.7 |
| G9 | 1.79 | 113.5 |
| G9 | 1.18 | 111.7 |
| Malbhog | 1.44 | 36.9 |
| Malbhog | 1.0 | 30.3 |
| Malbhog | 0.8 | 37.9 |
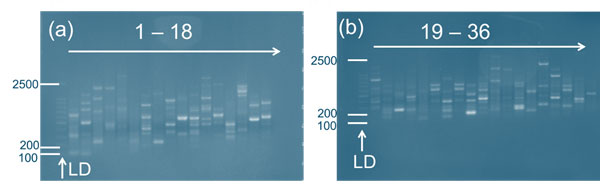
Analysis of Gel Electrophoresis: The results of agarose gel electrophoresis confirmed the successful extraction of high-molecular-weight DNA from both G9 and Malbhog banana cultivars. The genomic DNA amplification pattern of micropropagated Musa spp. (Malbhog and G9) using RAPD primers revealed distinct banding profiles, confirming the successful amplification of genomic DNA (Figures 1 and 2). The presence of intact DNA bands with minimal smearing indicated that the optimized CTAB-based extraction method effectively preserved DNA integrity. Additionally, the absence of RNA contamination suggested that the RNase treatment was efficient, ensuring the purity of the extracted DNA for downstream applications such as RAPD analysis and genetic fidelity assessments. Variations in band intensity were observed between the two cultivars despite the overall success of the DNA extraction process.
The DNA samples from the G9 cultivar exhibited stronger and more distinct bands, indicating a higher DNA yield and better integrity. In contrast, the Malbhog samples displayed fainter bands in some cases, suggesting lower DNA concentrations or partial degradation. This discrepancy could be attributed to differences in the biochemical composition of the cultivars, as Malbhog has a higher content of polysaccharides and phenolic compounds, which may interfere with DNA extraction and stability. The sequential use of primers 1–36, as listed in Table 1, ensured a comprehensive assessment of genomic stability and variation among the plantlets.
These findings highlight the importance of cultivar-specific optimization in DNA isolation protocols. While the current method performed well for G9, further refinements, such as additional purification steps or modifications in lysis conditions, may be necessary to enhance DNA yield and integrity for Malbhog. Ensuring high-quality DNA is crucial for genetic analyses, as degraded or impure DNA can affect the accuracy of PCR-based studies and other molecular applications.
Figure 1: Genomic DNA amplification pattern of micropropagated Musa spp. (G9) using RAPD primers. LD represents the DNA ladder. (a) Amplification with primers 1–18, (b) Amplification with primers 19–36, applied sequentially as listed in Table 1.
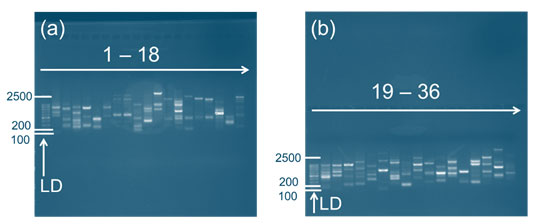
Figure 2: Genomic DNA amplification pattern of micropropagated Musa spp. (Malbhog) using RAPD primers. LD represents the DNA ladder. (a) Amplification with primers 1–18, (b) Amplification with primers 19–36, applied sequentially as listed in Table 1.
CONCLUSION
DNA was successfully extracted from G9 and Malbhog banana leaves using the CTAB technique. The variations in DNA yield and purity were observed, with G9 producing higher-quality DNA compared to Malbhog, which exhibited lower purity likely due to residual polysaccharides and phenolic compounds. While the extracted DNA was sufficient for genetic studies, further refinement of the extraction protocol is recommended, particularly for Malbhog, to enhance DNA purity and yield. Adjustments such as optimizing lysis conditions, incorporating additional purification steps, or modifying buffer compositions could improve overall extraction efficiency. Ensuring a reliable and reproducible DNA isolation method is crucial for accurate genetic fidelity assessments, RAPD analysis, and marker-assisted selection. These findings contribute to advancing molecular research on banana cultivars and support the development of efficient DNA extraction protocols for breeding and conservation efforts.
ACKNOWLEDGEMENTS
Authors thank the Head Department of Botany and Principal T.P.S. College, Patliputra University, and Head, Department of Botany, Patliputra University for their support and encouragement.
Conflict of Interest statement: The Authors declare no conflict of interest
Funding: Nil
Data Availability: Data are available with the corresponding author
REFERENCES
Aboul-Maaty, N.A.F. and Oraby, H.A.S., (2019). Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bulletin of the National Research Centre, 43(1), pp.1-10.
Chukwu, S.C., Awala, S.K., Angombe, S., Valombola, J.S., Nanhapo, P.I., Mberama, C., Rafii, M.Y., Oladosu, Y., Thomas, B., Okporie, E.O. and Musa, I., (2025). Recent progress in tissue culture techniques and biotechnological innovations for banana production (Musa spp.): a review. Discover Plants, 2(1), p.13.
Clarke, J.D., (2009). Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harbor Protocols, 2009(3), pp.pdb-prot5177.
Dhanapal, S., Sekar, D.S. and Satheesh, P.M., (2014). Efficiency of RAPD, SSR and ISSR markers in evaluating the genetic fidelity for micropropagated Musa accuminata plant exposed to coal extracted humic acid and commercially available products.
Emmanuel, J.K., Mtashobya, L.A. and Mgeni, S.T., (2025). Potential Contributions of Banana Fruits and Residues to Multiple Applications: An Overview. Natural Product Communications, 20(2), pp.1934578X251320151.
Evans, E.A., Ballen, F.H. and Siddiq, M., (2020). Banana production, global trade, consumption trends, postharvest handling, and processing. Handbook of banana production, postharvest science, processing technology, and nutrition, pp.1-18.
Horry, F.B.J. and Jenny, C., (2020). Making banana breeding more effective. In Achieving sustainable cultivation of bananas (pp. 217-256). Burleigh Dodds Science Publishing.
Jacobsen, K., Omondi, B.A., Almekinders, C., Alvarez, E., Blomme, G., Dita, M., Iskra‐Caruana, M.L., Ocimati, W., Tinzaara, W., Kumar, P.L. and Staver, C., (2019). Seed degeneration of banana planting materials: strategies for improved farmer access to healthy seed. Plant pathology, 68(2), pp.207-228.
Johari, S. and Majumder, S., (2015). An Efficient DNA extraction protocol for successful PCR detection of banana bunchy top virus from banana leaves. Asian Journal of Biotechnology, 7(2), pp.80-87.
Kumar, D., Chakradhar, P., Ranganna, G., Vimal, V.K., Raj, R., Anusha, C., Patra, S. and Nagaraju, V., (2024). Tissue culture in banana cultivation: A review of its impact on disease management, yield improvement, and sustainable production. Journal of Advances in Biology & Biotechnology, 27(9), pp.628-44.
Lázaro-Silva, D.N., De Mattos, J.C.P., Castro, H.C., Alves, G.G. and Amorim, L.M.F., (2015). The use of DNA extraction for molecular biology and biotechnology training: a practical and alternative approach. Creative Education, 6(08), p.762.
Ramesh, P., Mallikarjuna, G., Sameena, S., Kumar, A., Gurulakshmi, K., Reddy, B.V., Reddy, P.C.O. and Sekhar, A.C., (2020). Advancements in molecular marker technologies and their applications in diversity studies. Journal of Biosciences, 45, pp.1-15.
Sahin, E.C., Aydin, Y. and Uncuoglu, A.A., (2024). Molecular Marker Applications in the Selection of Elite Genotypes for Plant Stress Tolerance and Genetic Fidelity. OBM Genetics, 8(3), pp.1-25.
Santos, S.A., Félix, R., Pais, A.C., Rocha, S.M. and Silvestre, A.J., (2019). The quest for phenolic compounds from macroalgae: A review of extraction and identification methodologies. Biomolecules, 9(12), p.847.
Satyanarayana, S.D., Krishna, M.S.R. and Kumar, P.P., (2017). Optimization of high-yielding protocol for DNA extraction from the forest rhizosphere microbes. 3 Biotech, 7, pp.1-9.
Smith, M.K., (1988). A review of factors influencing the genetic stability of micropropagated bananas. Fruits, 43(4), pp.219-223.
Spadoni, A., Sion, S., Gadaleta, S., Savoia, M.A., Piarulli, L., Fanelli, V., Di Rienzo, V., Taranto, F., Miazzi, M.M., Montemurro, C. and Sabetta, W., (2019). A simple and rapid method for genomic DNA extraction and microsatellite analysis in tree plants. Journal of Agricultural Science and Technology, 21(5), pp.1215-1226.
Suman, S., (2017). Plant tissue culture: A promising tool of quality material production with special reference to micropropagation of banana. Biochemical & Cellular Archives, 17(1).
Susantini, E., Lisdiana, L., Isnawati, Tanzih Al Haq, A. and Trimulyono, G., (2017). Designing easy DNA extraction: teaching creativity through laboratory practice. Biochemistry and Molecular Biology Education, 45(3), pp.216-225.
Turaki, A.A., Ahmad, B., Magaji, U.F., Abdulrazak, U.K., Yusuf, B.A. and Hamza, A.B., (2017). Optimised cetyltrimethylammonium bromide (CTAB) DNA extraction method of plant leaf with high polysaccharide and polyphenolic compounds for downstream reliable molecular analyses. African Journal of Biotechnology, 16(24), pp.1354-1365.
Vinod, K.K., (2004). Total genomic DNA extraction, quality check and quantitation. Tamil Nadu Agricultural University, Coimbatore, pp.109-121.
Xin, Z. and Chen, J., (2012). A high throughput DNA extraction method with high yield and quality. Plant methods, 8, pp.1-7.


