1Animal Health Division, Central Institute for Research on Goats, Makhdoom,
Farah, Mathura, Uttar Pradesh, India.
2Departments of Biotechnology, GLA University, Mathura, Uttar Pradesh, India.
Corresponding author email: geetikagupta04@gmail.com
Article Publishing History
Received: 21/10/2021
Accepted After Revision: 19/12/2021
Globally antibiotic resistance has become a major concern, which warrants the real time monitoring for resistance in very common pathogenic organisms. E. coli is normal micro flora in humans, but sometimes it can be pathogenic. For the observation and increment of antimicrobial resistance among pathogen, E. coli has been one of the important pathogens. It is present everywhere in fecal, water, food etc., if resistant E. coli will present in the environment that it can be transferrable anywhere through water, fecal food, animals and humans. This is very dangerous to living beings. This study was designed on status of antibiotic resistance in E. coli isolates in human kids and animal kids, both.
Newborns are affected more because of poor or lack of immune system. In this study, fecal materials were used as sample material collected from goat kids (0-3 months) and human children (up to 3 years) residing in same local area. Fifteen fecal samples were collected from human children (up to 3 years) and goat kids (0-3 months) in each case to study the risk of transmission of resistance in E. coli isolates. PCR was conducted on genomic DNA isolates for the presence of usp A gene of E. coli. Multiplex PCR were conducted on plasmid DNA isolates for the resistance specific genes.
Molecular resistance results in goat kids isolates showed resistance to antibiotics with tetracycline, sulphonamide, gentamycin, streptomycin and cephalothin to the level of 93.33, 53.33, 46.66, 13.33 & 6.66% respectively, whereas, human E. coli isolates were showed the highest resistance to sulphonamide, Tetracycline and β-lactams were as 53.33, 46.66 and 13.33% respectively but no resistance with gentamycin and streptomycin. Here, we concluded that humans and animals both were refractory to the various groups of antibiotics. This study will help in making the strategy for prevention or reduction of resistance in public
Coli, Genomic DNA, Plasmid DNA, Resistance.
Gupta G, Kumar A, Bhardwaj A. Comparative Study of Plasmid-Mediated Molecular Resistance Status of Diarrheic Escherichia coli Isolate in Human and Goat Kids. Biosc.Biotech.Res.Comm. 2021;14(4).
Gupta G, Kumar A, Bhardwaj A. Comparative Study of Plasmid-Mediated Molecular Resistance Status of Diarrheic Escherichia coli Isolate in Human and Goat Kids. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/3FejWHl“>https://bit.ly/3FejWHl</a>
Copyright © Gupta et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Childhood diarrhea is a major public health problem and second leading cause of high mortality below 5 year of age children’s. Nearly 1.7 billion cases of childhood diarrheal diseases have been reported every year, killed around 525,000 five year age children, rating for 8% of all deaths worldwide. The deaths from diarrhea mostly occur in children below 2 years of age. Diarrheal diseases have a detrimental impact on child growth and cognitive development. Increased risks of malnutrition in children are associated with diarrheal diseases. The prevalence of the disease remains at an alarming rate where infants and children are unmoving at risk of death and other complications, while over the past two decades the decrease in the episodes of childhood diarrhea is observable globally (Paul 2020).
- coli is Gram-negative, non-spore forming, facultative anaerobic bacteria belongs to the Enterobacteriaceae family. Natural gastrointestinal flora part of the humans and warm blooded animals forms from it (Aijuka and Buys 2019). E. coli association in animals such as goats has major significance for infection in humans, particularly rearing goats as backyard farming. Infection to humans can transmit through animals as their infected and diseased meat or between handling procedures or through ingestion by the consumer (Zerabruk et al. 2019; Paul 2020).
Also, during slaughtering via fecal contamination, E. coli presence in animal feces allows the entry in the food chain resistant bacteria to antibiotics is present everywhere in surroundings and greatly increased their obstructive effect. Most E. coli are harmless commensal organisms (Zerabruk et al. 2019). E. coli are an opportunistic pathogen that can endure well in aquatic systems and are exceptionally proficient at horizontal gene transfer, which is believed to be the vector for antibiotic-resistance dissemination (Maeusli et al. 2020). The study was aimed to molecular resistance typing of E. coli isolates both in young age human children and in goat kids (1) Frequency of single resistant gene in both (2) Frequency of one or more resistant gene (MDR) in both and (3) which group of antibiotics was showed more resistance.
MATERIAL AND METHODS
Fecal samples were collected in goat kid’s belonging to 0-3 month age group from rectum with the help of cotton swab (HI media, India) and from human child up to 3 years directly without touching the surface. Human samples were collected with the consent of parents/guardians. After collection samples were directly stored in the ice box for transportation. If possible, fresh samples were processed for identification in the laboratory or if not possible then immediately samples were stored at -70° C until the use.
For identification of bacteria, fecal samples were directly streaked on enriched media brain heart infusion solid media for growth from which a single colony used for further experiments such as lactose fermentation on differential media, streaking on selective media (eosin methylene blue media) for confirmation. To hold E. coli isolates for further process, all isolates were maintained and stored at a temperature of -70°C. Staining was performed from gram staining kit (BD & BBL Difco, USA) as per the method described in the previous studies (Hedge et al. 2013). Biochemical tests were performed from the Himedia kit (HI media, Mumbai, India) and Sigma kits (Japan). DNA from a cultural broth was isolated as per Yang et al., 2008. Isolated DNA was stored at -70° C until their use.
Polymerase chain reaction (PCR) test was performed for the usp A gene of E. coli in Peq lab thermo cycler. The reaction mixture (25µl) were prepared as 22.5 µl Invitrogen master mix (accuprime), 1µl DNA template, and .75 µl of each primer forward and reverse. PCR conditions were as 95°C (5 minute); 30 cycles of 95°C (1 minute), 58 °C (30 seconds) 72°C (1 minute); and a final extension at 72°C (5 minutes) stored at 4°C for infinite. PCR product was seen in the Gel doc machine with the help of gel electrophoresis. 1.3 % agarose gel containing ethidium bromide was prepared in a casting tray. After solidification, PCR products were filled in the well with 6x gel loading dye (Thermo scientific), then run on the power supply at 60 volts for 1 hour in a gel tank filled with TAE buffer.
Fifteen human and fifteen goat kids E. coli isolates processed for Plasmid DNA isolation whereas, a single colony was inoculated in Luria bertani broth and incubate at 37°C for 3-4 hour. After incubation culture broth centrifuge at 1500 rpm for 5 minutes and supernatant discarded. Pellet was washed with PBS 2-3 times and processed for DNA isolation as per the GSure Mini Kit protocol (GCC, New Delhi). IsolatedNA was stored in-70° C until the use. Polymerase chain reaction (PCR) tests were performed in Peqstar 96 x universal gradient thermocycler (PEQLAB Biotechnologie GmbH, Germany).
The reaction mixture (25µl) was prepared as 10µl emerald master mix (Takara, New Delhi), 1 µl DNA template and 1 µl of each primer forward and reverse and the remaining amount (12 µl) of nuclease – free water. PCR conditions for Cep were as 95°C for 5 min.;30 cycles of 95°C for 1 min, 55°C for 30 sec respectively72°C for 1 min.; and a final extension at 72°C for 5 min stored at 4°C for infinite time. PCR products were seen in the gel doc machine with the help of gel electrophoresis.1.3 % agarose gel was prepared in a casting tray.
After solidification, PCR products were filled in the well with 6x gel loading dye (Thermo Scientific, US). Run on the power supply at 60 volts for 1 hour in the gel tank filled with TAE buffer. For multiplex PCR, the reaction mixture was prepared as listed above, and in reaction mixture added all four genes forward and reverse primer in the same tube and decrease the amount of water.PCR conditions for (Gentamycin- Gen), (Tetracycline – Tet), (Sulphonamide – Sul) and (Streptomycin – Strp) were as 95°C (5 minute); 30 cycles of 95°C (1 minute), 53.5°C (30 second) respectively72°C (1 minute); and final extension at 72°C (5 minute) stored at 4°C for an infinite time. Results were seen as per the previous method.
RESULTS AND DISCUSSION
E. coli were characterized based on cultural, biochemical, and molecular methods in the laboratory. Observed findings on bacteria were as gram – negative and rod – shaped (Fig.1),
Figure 1: Microscopic result showing pink colour (gram-ve) rod shaped bacteria (E. coli)
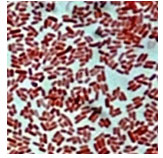
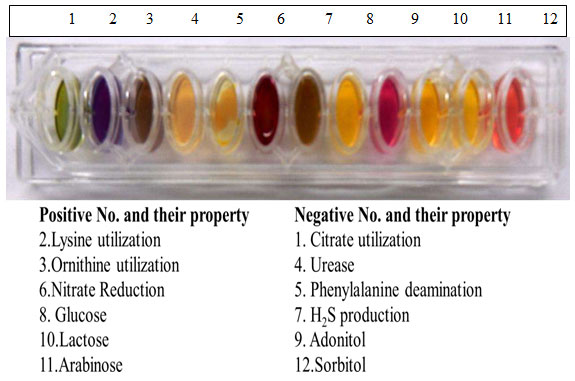
Figure 2: Bacteria showing biochemical Test positive or negative
Pale yellow colonies on Brain heart infusion agar, smooth pink colonies on Mac Conkey agar and green metallic sheen on EMB were observed. (Fig. 3, 4 & 5).
Figure 3: E. coli showing pale yellow colonies on BHI media
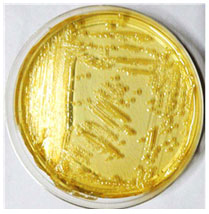
Figure 4: E. coli showing lactose fermentation on MCA media
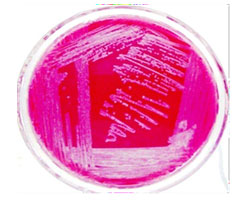
Figure 5: E. coli showing metallic sheen on EMB Media
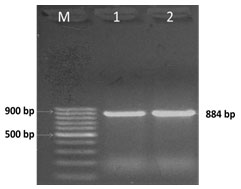
E. coli was confirmed at 884 bp as a single amplicon (Fig. 6) (Mishra et al. 2019).
Figure 6: PCR amplification of uspA gene; Lane M-DNA Marker, Lane 1- Positive
control, Lane 2-Positive PCR product
The primers were used in the study for confirmation and resistance as described in the previous studies respectively procured from Eurofins (Table-1) (Momtaz et al. 2012; Mishra et al. 2019).
Table 1. The primers used in the study for molecular and resistance gene confirmation of E. coli
| Primer (Gene) | Primer sequence | Product length (bp) |
|
UPEC (usp A) |
(F)CCGATACGCTGCCAATCAGT
(R)ACGCAGACCGTAGGCCAGAT |
884 |
| β-lactams (Cep) | (F)TGGCCAGAACTGACAGGCAAA
(R)TTTCTCCTGAACGTGGCTGGC |
462 |
| Gentamycin (Gen) | (F)CTTCAGGATGGCAAGTTGGT
(R)TCATCTCGTTCTCCGCTCAT |
286 |
| Streptomycin (Strp) | (F)TATCCAGCTAAGCGCGAACT
(R)ATTTGCCGACTACCTTGGTC |
447 |
| Tetracycline (Tet) | (F)GGTTCACTCGAACGACGTCA
(R)CTGTCCGACAAGTTGCATGA |
577 |
| Sulphonamide (Sul) | (F)TTCGGCATTCTGAATCTCAC
(R) ATGATCTAACCCTCGGTCTC |
822 |
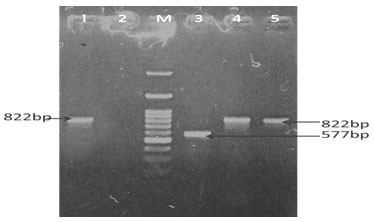
Different genes such as gen, strp, cep, tet, and sul resistant genes from the E. coli were successfully amplified using species – specific primers in animals and as well as in humans. PCR Amplification resulted in a single amplicon of 286,447,462,577& 822-bp as illustrated (Fig. 7, 8, 9 & 10) (Momtaz et al. 2012; Rubab and Oh 2021).
Figure 7: PCR amplification of strp, sul and tet gene; Lane M-DNA Marker Lane1 –Positive
PCR product (strp and tet) Lane 2-Positive PCR product (sul)
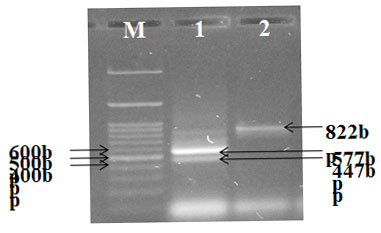
Figure 8: PCR amplification of cep gene; Lane M-DNA Marker Lane1, 2 & 4 –Positive
PCR product (cep) Lane 3-Negative PCR product (cep)
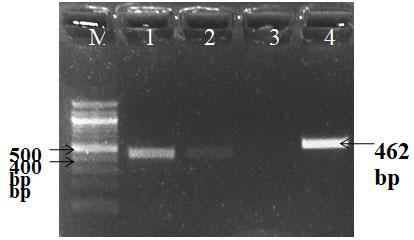
Figure 9: PCR amplification of Gen gene; Lane M-DNA Marker Lane 1-Negative PCR product,
Lane2-Negative PCR product, Lane3-Positive PCR product Lane4- Positive control
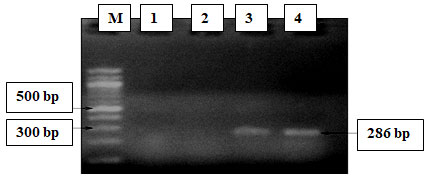
Figure 10: PCR amplification of sul and tet gene; Lane M-DNA Marker Lane 1 -Positive control (Sul), Lane2-Negative control (sul), Lane 3-Positive PCR product (tet) Lane 4&5–Positive PCR product
In our results, it was observed that genes associated with resistance to gentamycin, streptomycin sulphonamide, cephalothin, and tetracycline observed in 46.66, 13.33, 53.33, 6.66%, and 93.33% samples respectively in goat kids, Tetracycline, cephalothin, and sulphonamide resistance genes were observed as 26.66, 13.33, and 53.33% respectively in human kid samples and no gene resistance to gentamycin and streptomycin was recorded in humans’ samples (Table 2, 3 & 4 and graph 1, 2 &3).
Table2 .Frequency of resistant antibiotic genes was present in goat kids and human kids.
|
Group of antibiotics |
Gene |
No. of tested |
No. of resistant isolates (Percentage) | |
| Goat kids | Human kids | |||
| Aminoglycoside | Gen |
15-15 each |
7 (46.66) | 0 (0) |
| Strp | 2 (13.33) | 0(0) | ||
| Sulphonamide | Sul | 8 (53.33) | 8 (53.33) | |
| β-lactams | Cep | 1 (6.66) | 2 (13.33) | |
| Tetracycline | Tet | 14 (93.33) | 4 (26.66) | |
Graph 1
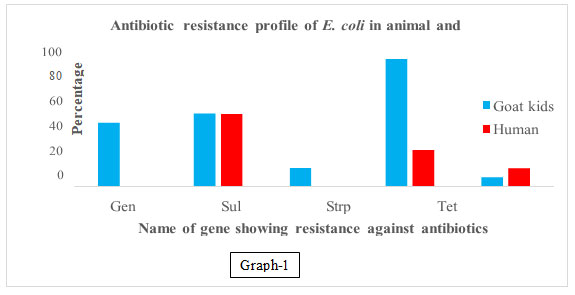
Table 3. Frequency of multi drug resistant antibiotic genes was present
in goat kids and young age children’s.
| Resistant to Genes | No. of tested | No. of resistant isolates (Percentage) | |
| Goat kids | Human kids | ||
| Only 1 |
15-15 each |
5 (33.33) | 7 (46.66) |
| 2 | 5(33.33) | 2 (13.33) | |
| 3 | 3 (20.00) | 1 (6.66) | |
| 4 | 2 (13.33) | 0(0) | |
| 5 | 0 (0) | 0(0) | |
Graph 2
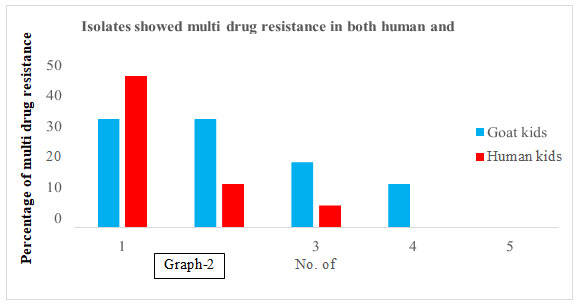
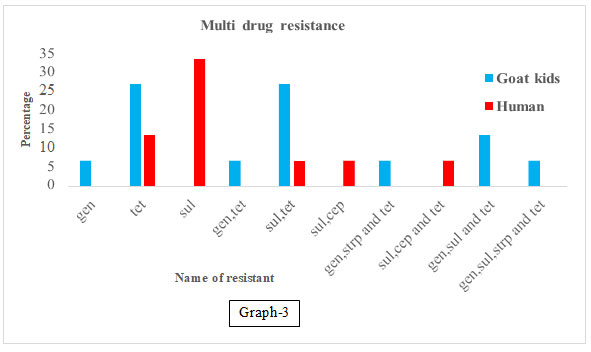
Table 4. Name of multi drug resistant antibiotic genes was present
in goat kids and young age children’s.
| Group of antibiotics | Name of exactly resistant Genes | No.of tested | No. of resistant isolates (%) | |
| Goat kids | Human kids | |||
| Aminoglycoside (AG) | Gen | (15-15) | 1 (6.66) | 0 |
| Tetracycline (T) | Tet | 4(26.66) | 2 (13.33) | |
| Sulphonamide (Sulk) | Sul | 0 | 5 | |
| AG and Tetracycline | Gen, Tet | 1(6.66) | 0 | |
| Sul and T | Sul, Tet | 4 (26.66) | 1(6.66) | |
| Sul and β-lactams | Sul, Cep | 0 | 1(6.66) | |
| AG and Tetracycline | Gen, Strp
and Tet |
1(6.66) | 0 | |
| Sul, β-lactams and Tetracycline | Sul, Cep
and Tet |
0 | 1(6.66) | |
| AG, Sulphonamide and Tetracycline | Gen, Sul
and Tet |
2(13.33) | 0 | |
| AG, Sul, β-lactams and Tetracycline | Gen, Sul, Cep
and Tet |
1(6.66) | 0 | |
| AG, Sulphonamide and Tetracycline | Gen, Sul, Strp and Tet | 1(6.66) | 0 | |
Graph 3
Our results were similar in goat kids except for streptomycin where a high prevalence of tetracycline (36 to 97%), sulfathiazole (50 to 100%), and streptomycin (53 to 100%) resistance was detected in chickens with similar to previous studies (Smith et al. 2007). In a study have reported that tetracycline and sulphonamide resistance in E. coli from slaughtered commercial chickens were 52.6, 47.36 % respectively. They also reported amino glycosides and β- lactams resistance in chickens were nil (Momtaz et al. 2012; Rubab and Oh 2021).
In study, frequency of streptomycin, tetracycline, gentamycin, and sulphonamide genes were in 96.10%, 85.06%, 54.54%, and 40.25%, respectively in E. coli from pediatric patients, theariation in this study may be attributed to the level of exposure to antibiotics in goat kid (Heidary et al. 2013). ported allow frequency of occurrence of genes of tetracycline, aminoglycosides and other β-lactamases genes perform well as signs of emerging resistance in children, unlike the sulphonamide resistance (Singh et al. 2019). The study reported tetracycline, sul1 (for sulphonamide), streptomycin resistance followed by 77.17%, 45.94% and 34.65% from E coli isolate from chicken meat (Rahman et al. 2020).
In a study reported that STEC isolates from different sources show the highest presence of ampC genes with a frequency of 47%. The detection rates tet (A) were 35%, respectively. Nearly 74.5% of those isolates were resistant to all of the tested antibiotic resistance genes. In our results, in case of goat kid 100 percent while in case of human kids 33.33% E. coli isolate were resistant to all of the tested antibiotic resistance genes (Rubab and Oh 2021). In our results 26.66 % E. coli isolates form goat kids were showed multi drug resistance to group of antibiotics as Aminoglycoside, Sulphonamide and Tetracycline.
CONCLUSION
The findings of the present study suggests that the antibiotic resistance is observed in E. coli isolated from both humans and goat kids. Sulphonamide group of antibiotics showed resistance in both. In goat kids, tetracycline showed the highest resistance and sulphonamide in humans. In our study observed that E. coli isolated from goat kids showed resistance to amino glycoside, tetracycline, β – lactams, and sulphonamide group of antibiotics but E. coli isolated from human kids showed resistance to tetracycline, β –lactams, and sulphonamide group of antibiotics. Goat isolates showed resistance to four groups of antibiotics, while human isolate showed resistance to three groups of antibiotics.
ACKNOWLEDGEMENTS
This work was financially supported by Indian Council of Agriculture Research, New Delhi under AINP-NM Project.
Conflict of Interests: Authors declare no conflict of interests to disclose.
REFERENCES
Aijuka M. and. Buys E. M (2019). Persistence of food borne diarrheagenic Escherichia coli in the agricultural and food production environment: implications for food safety and public health. Food Microbiology, Vol 82 pp 363–370.
Hegde R, Isloor S, Prabhu KN, et al. (2013). Incidence of subclinical mastitis and prevalence of major mastitis pathogens in organized farms and unorganized sectors. Indian Journal Microbiology, Vol 53(3) pp 315-320.
Heidary M, Momtaz H and Madani M (2014). Characterization of Diarrheagenic Antimicrobial Resistant Escherichia coli Isolated From Pediatric Patients in Tehran, Iran. Iran Red Crescent Med J., Vol 16(4): e12329. DOI:10.5812/ircmj.12329.
Maeusli M, Lee B, Miller S, et al. (2020). Horizontal gene transfer of antibiotic resistance from Acinetobacter baylyi to Escherichia coli on lettuce and subsequent antibiotic resistance transmission to the gut microbiome. mSphere, Vol 5 (3), e00329-20.
Mishra AK, Singh DD, Gururaj K, et al. (2019). Molecular Characterization of Diarrhoegenic Escherichia coli Isolated from Neonatal Goat-Kids. Journal of Animal Research Vol 9 (1) pp 51-59.
Momtaz H, Rahimi E and Moshkelani S (2012). Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Veterinary Medicina, Vol 57 (4) pp 193–197.
Paul P (2020). Socio-demographic and environmental factors associated with diarrheal disease among children under five in India. BMC Public Health, 20:1886. https://doi.org/10.1186/s12889-020-09981-y.
Singh T, Singh PK, Dar SA, et al. (2019). Changing paradigm of antibiotic resistance amongst Escherichia coli isolates in Indian pediatric population. PloS one, Vol 14(4): e0213850.https://doi.org/10.1371/journal.pone.0213850.
Smith J L, Drum D J V, Dai Y, et al. (2007). Impact of Antimicrobial Usage on Antimicrobial Resistance in Commensal Escherichia coli Strains Colonizing Broiler Chickens. Applied and environmental microbiology, Vol 73(5) pp 1404–1414.
Yang JL, Wang MS, Cheng AC, et al. (2008). A simple and rapid method for extracting bacterial DNA from intestinal micro flora for ERIC-PCR detection. World Journal Gastroenterology, Vol 14(18) pp 2872-2876.
Zerabruk K., Retta N, Muleta D, et al. (2019). Assessment of microbiological safety and quality of minced meat and meat contact surfaces in selected butcher shops of Addis Ababa, Ethiopia. Journal of Food Quality, Vol 2019 (53) pp1-9.


