Faculty of Biosciences, Institute of Biosciences and Technology, Shri
Ramswaroop Memorial University, Lucknow, Uttar Pradesh, India.
Corresponding author email: neeraj.bio@srmu.ac.in
Article Publishing History
Received: 29/06/2021
Accepted After Revision: 18/09/2021
Laccases are biotechnologically relevant enzymes used in different industries including textile industry, food processing industry, pharmaceutical industry, wood processing industry and chemical industry. Recently they are used in biosensors for detection of phenolic compounds, in biofuel cells for the generation of eco-friendly energy and also as a medical diagnostics tool. Because of wide range of applications, laccases have received so much of attention of scientists and researchers in the last few decades. They are extracellular, metalloenzymes containing copper.
It has ability to catalyses the oxidation of polyphenolic compounds. Laccase activity was reported in different range of fungi including white-rot fungi basidiomycetes, deuteromycetes and ascomycetes where its main role is lignin-degrading. The searching of new microbial sources for laccase production is great demand. Present study was focused to search a new microbial strain that produces laccase from different soil samples. The strains isolated from soil enabled the laccase production and was qualitatively analyzed using 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonate) (ABTS). It is an indicator compound used for the rapid visual of laccase positives strain. Laccase isolates were identified with the change in color from dark green to purple on ABTS containing Potato dextrose agar (PDA) plates.
The fungal isolates were identified by conventional methods and molecular characterization techniques using DNA extraction, polymerase chain reaction (PCR) amplification and sequence analysis. Genomic DNA was isolated, internal transcribed spacer ribosomal DNA (ITS-rDNA) amplified by PCR and ITS regions were sequenced by Sanger sequencing method. Based on conventional methods and molecular characterization, the laccase producing isolate was identified as Alternaria alternata LF9_CPD_NRRI MK855476.1. Genetic distance and neighbor-joining algorithm was analyzed by using MEGA 6.0 software and phylogenetic tree was constructed to study the evolutionary history.
ABTS, Laccase, Lignocellulolytic, Oxidoreductase, White Root Fungus,
Singh D, Gupta N. Screening and Molecular Characterization of Ligninolytic Enzyme-Producing Strain Alternaria alternata from soil. Biosc.Biotech.Res.Comm. 2021;14(3).
Singh D, Gupta N. Screening and Molecular Characterization of Ligninolytic Enzyme-Producing Strain Alternaria alternata from soil. Biosc.Biotech.Res.Comm. 2021;14(3). Available from: <a href=”https://bit.ly/3xLw6nf“>https://bit.ly/3xLw6nf</a>
Copyright © Singh and Gupta This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Laccases [benzenediol: oxidoreductase, enzyme commission (EC) 1.10.3.2] are most promising lignin-degrading enzymes also called lignocelluloses. These multifunctional enzymes oxidize phenolic compounds, including aromatic amines, by reducing molecular oxygen into water. Laccase possesses a wide-range of substrate specificity also known as extracellular globular proteins with 50-130 kilo-Dalton (kDa) molecular weight. Optimum pH at which laccase shows maximum activity is below 7, but a few studies in the past have reported that laccase can work at higher pH (Viswanath et al. 2014; Teerapatsakul et al. 2016).
Because of their significant utilization in various biological processes, the enormous demand of laccases is increasing day by day in many industrial fields of bioremediation and xenobiotic degradation, dye degradation, biosensor, and food and bakery. Various improvements are undergoing to better the formation of lignocellulose-based products in nanotechnology application (Margot et al. 2013; Rao et al. 2019).
Laccases are widespread in nature and was first reported from Rhus vernicifera in Japanese lacquer tree. Laccase is obtained from wide variety of organisms including bacteria to higher organism such as fungi, plants and insects. Laccases expressed by such microorganisms includes majorly lignin peroxidase (LiP), manganese peroxidase (MnP) and laccases (Dwivedi et al. 2011; Xu et al. 2017).
Several prominent lignin-degrading enzymes have been classified from fungal groups and those belonging to ascomycetes, basidiomycetes and deuteromycetes (Gedikli et al. 2010). Basidiomycetes are great producers of laccase enzyme such as Cerena maxima, Coriolopsis polyzona, Lentinus tigrinus, Trametes versicolor, Trametes hirsute, Trametes ochracea, Trametes villosa, Pycnoporus sanguineus, and Pleurotus eryngii etc (Shleev et al. 2004; Gedikli et al. 2010; Deepa et al. 2020).
The laccase producing fungi isolated from new microbial systems would be considered, extremely useful for achieving several biotechnological applications. These fungal laccases are very efficient in conversion of plastic, fuel, paint and wood components into nutrients. Currently researchers are mainly focusing on enhancing laccase production from several strains of Basidiomycetes predominantly wood-root-fungi by optimizing culture conditions suitable for production. Couple of studies have reported ascomycetes on their lignin degrading ability but very less reported on Alternaria alternate for laccase production.
Recently laccase was reported in Bacillus species isolated from industrial waste (Glaeser et al. 2010; Gassara et al. 2010; Yadav et al. 2019; Deepa et al. 2020). Fungi known as biological controllers illustrates the huge fungal diversity and some vital fungal groups are identified at global extent (Delgado-Baquerizo et al. 2017; Irfan et al. 2018). The objectives of the present work were to isolate laccase producing fungi from microbial system in natural habitat, screen for the presence of lignin-modifying enzyme using specific substrate containing ABTS 2,2’-azinobis-(3 ethylbenzthiazoline-6-sulfonate) as color indicator compound to identify potential fungal strains that are able to produce laccase using molecular analysis and construct phylogenetic tree.
MATERIAL AND METHODS
ABTS (2, 2′-azino-bis (3-ethylbenzothiazoline-6-sulfonicacid) was purchased from Hi-Media Pvt. Ltd Lucknow, India. Other chemicals used were of analytical grade and were purchased from local markets. Soil samples were collected from the campus of Shri Ramswaroop Memorial University, Lucknow Deva-road, Barabanki, Uttar Pradesh, India located at approximately 123 meters (404 ft) above sea level coordinates: latitude: 26°50′21″N and longitude: 80°55′23″ E. Moderate rain fall region with the temperature ranges maximum 47.50C to 2.50C minimum (Hannula et al., 2017).
The sample collecting site covered with various trees and shrubs, turnover of the organic substance in such soil expresses rapidly. The soil sample was collected in sterile plastic bags aseptically from a depth of 10–15 cm below the earth’s surface as shown in Fig. 1.
Figure 1: Site for the collection of soil sample (Source: Google map, https://goo.gl/maps/kHkWMFV362NoDcGD6)

For the isolation of microbes (ascomycetes and fungi), the soil samples were first serially diluted and then were spread evenly over the surface of potato dextrose agar plates to isolate fungi (Kanaujia et al. 2014). Further, the plates were incubated at 35oC and monitored after 5th days. Fungal screening was carried out by using ABTS 2,2’-azinobis-(3 ethylbenzthiazoline-6-sulfonate) as indicator compound.
Positive laccase isolates were visualized as dark green to purple color on ABTS containing Potato dextrose agar (PDA) plates which indicates the presence of laccase being produced by the fungus, as previously reported (Pointing et al., 2000). Screening of laccase producing organisms was employed on PDA plates supplemented with 0.1% and 0.01% ABTS 2,2’-azinobis-(3 ethylbenzthiazoline-6-sulfonate) respectively and incubated at 30OC for 7 days. Culture plates indicate that the definite color changes were considered for laccase producing strain and employed further for consequent studies (Pointing et al., 2000).
Total genomic DNA was extracted from the fungal strain by genomic DNA extraction kit method. Comparison of the internal transcribed spacer (ITS) ribosomal DNA (rDNA) gene sequence was employed for molecular characterization. The gene encoding ITS region was amplified by the polymerase chain reaction (PCR) using forward (CTGGTGCCAGCAGCCGCGGYAA) and reverse primer (CKRAGGGCATYACWGACCTGTTAT3).
All PCR amplifications were carried out in a Thermal Cycler (Model No. 9902). A hot-start procedure (94 °C for 3 min) was performed and enzyme was added earlier to avoid non-specific annealing of primers. DNA was amplified during 30 cycles of 94 °C for 30 sec, 48 °C for 30 sec, 72 °C for 1 min and 72 °C for 7 min.
After purification of amplified products by agarose gel electrophoresis, the amplified products were sent for sequencing to Xcelris Labs Limited Ahmedabad, India. The most homologous sequence was determined by comparison to Gene Bank database using BLAST software. Genetic distance and neighbor-joining algorithm was analyzed by using MEGA 6.0 software and the phylogenetic tree was constructed to study the evolutionary relation between species.
RESULTS AND DISCUSSION
SCREENING FOR LACCASE ACTIVITY USING ABTS ASSAY: The main purpose of screening is to select fungi with desired characteristics proposed for miscellaneous applications of food industries, paper and pulp industry, bioremediation of environmental pollutants, textile industries, biosensor application, organic synthesis and biofuel formation, bioremediation of toxic chemical wastes, pharmaceutical and cosmetics industries, and nanobiotechnology applications (Devasia et al. 2016; Zeng et al. 2017; Zdarta et al. 2018; Veraet al. 2019).
All the isolated and subculture fungal colonies were inoculated on PDA plates supplemented with specific substrate for laccase. Five days old culture was placed on PDA plates containing ABTS and incubated at 30°C for 7 days (Antecka et al. 2018).
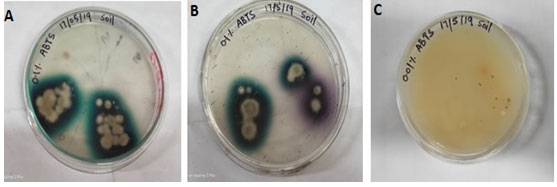
After 8th days the culture plate was able to develop dark green color on 0.1% ABTS 2,2’-azinobis-(3 ethylbenzthiazoline-6-sulfonate) containing PDA plate as shown in (Fig. 2. A) and purple color on 0.1% ABTS containing PDA medium petri-plate indicates the presence of laccase being produced by the fungus as shown in Fig. 2. B (Vantamuri and Kaliwal 2015). The 0.01% ABTS containing PDA plate shows no reaction as shown in Fig. 2. C.
Figure 2: (A) Screening of Laccase producing Alternaria alternate isolate LF9_CPD_NRRI on PDA plates containing 0.1% ABTS substrate showing intense dark green color halos around the colony. (B) PDA plate containing the 0.1% ABTS substrates showing intense dark green to purple color halo around the colony. (C) PDA plates containing the 0.01% ABTS substrates showing no reaction.
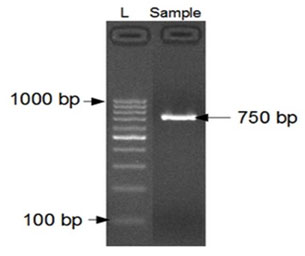
PCR AMPLIFICATION AND IDENTIFICATION OF THE FUNGAL STRAIN: The amplified DNA quality was evaluated on 1.2% agarose gel, a single discrete high-molecular weight PCR amplicon band of 750 bp has been observed as shown in Fig. 3.
Figure 3: 1.2% Agarose gel showing 1000bp amplicon (SSU region) of 18S rDNA.
Lane 1: 1000bp DNA Ladder and Lane 2: 750bp amplicon (SSU region) of 18S rDNA.
SEQUENCE ANALYSIS: Consensus sequence of 750 bp of 18S gene in SSU region was generated from forward and reverse sequence data using aligner software. The 18S gene in SSU region sequence was used to carry out BLAST alignment search tool of NCBI GenBank database. Based on maximum identity score first fifteen sequences were selected and aligned using multiple alignment software program Clustal W. Distance matrix was generated using RDP database and the phylogenetic tree was constructed using MEGA 6.0 as shown in Fig. 4.
Figure 4: Evolutionary Relationship
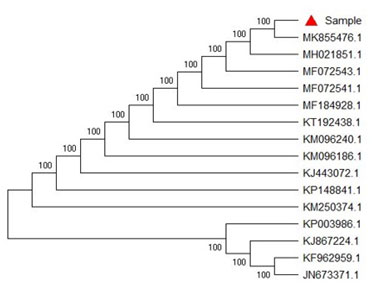
Table 1. Alignment view table showing sequences producing significant alignments from NCBI GenBank
| Accession | Description | Max score | Total score | Query coverage | E-value | Max ident |
| MK855476.1 | Alternaria alternataLF9_CPD_NRRI | 1424 | 1424 | 100% | 0.0 | 99.49% |
| MH021851.1 | Alternaria alternata isolate PCPDI | 1424 | 1424 | 100% | 0.0 | 99.49% |
| MF072543.1 | Alternaria alternata strain 013 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| MF072541.1 | Alternaria alternata strain 003 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| MF184928.1 | Alternaria sp. Isolate QDFBLJ-1 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KT192438.1 | Alternaria sp.PMK2 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KM096240.1 | Alternaria sp.MF382 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KM096186.1 | Alternaria sp. LF255 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KJ443072.1 | Alternaria alternata strain G408 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KP148841.1 | Fungal sp.S1 ZM-2014 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KM250374.1 | Alternaria sp. SPS-04 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KP003986.1 | Alternaria alternata strain J14 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KJ867224.1 | Alternaria sp. DBC-AD | 1424 | 1424 | 100% | 0.0 | 99.49% |
| KF962959.1 | Alternaria alternata strain HA4087 | 1424 | 1424 | 100% | 0.0 | 99.49% |
| JN67337.1 | Alternaria alternata strain HDJZ-zwm-34 | 1424 | 1424 | 100% | 0.0 | 99.49% |
The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed (Saitou et al. 1987; Phatake et al. 2015).
Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches. The evolutionary distances were computed using the Kimura 2-parameter method and were in the units of the number of base substitutions per site (Kimura 1980; Felsenstein 1985).
This analysis involved 16 nucleotide sequences. Codon positions included were 1st+2nd+3rd+non-coding. All ambiguous positions were removed for each sequence pair (pair wise deletion option). There was a total of 785 positions in the final dataset. Evolutionary analyses were conducted in MEGA 6.0 (Tamura et al. 2011; Phatake et al. 2015; Senthivelan et al. 2019).
The extracellular laccase producing fungus was isolated from a new habitat and used for enzyme production. Based on the screening results using lignolytic substrates of ABTS and identified by dark green and purple color oxidation zone was developed around the colonies (positive ABTS-substrate oxidation) which were taken for the laccase production (Wang et al. 2018; Senthivelan et al. 2019). Depending upon the observations one predominant strain of fungus was selected for further study. The screening results showed the formation of color under the medium with ABTS (substrate) confirmed that the isolated fungus has ability to produce lignin-degrading enzyme (Das et al. 2016).
The newly isolated fungal strain was further subjected to their morphological analysis. By virtue of their morphological identification, the isolated fungus belonged to the class of Alternaria fungal species. Further the fungus was identified as Alternaria alternata isolate LF9_CPD_NRRI (Accession Number: MK855476.1) by molecular identification of 18s rRNA sequencing method. The morphological characteristics similarly reported previously; the colonies of Alternaria fungus were olive-black in color with white margins (Abeer et al. 2015; Sharma et al. 2016; Thakkar et al. 2020).
Conidiophore septum sometimes demonstrated as branched or simple straight or curved shaped medium brown and sleek barred, Conidia like branched chains, short conical or cylindrical beak and they had been obscured in many previous studies (Abeer et al. 2015; Sharma et al. 2016). Similar investigations were also reported in the ABTS containing plate showed the green and purple-color in the presence of laccase producing fungi defined the positive reaction for the laccase production (Vantamuri and Kaliwal 2015).
The efficiency of laccase producing strain Alternaria alternata was reported at 30OC on 7thday of incubation period (Tapwal et al., 2014). Recently new laccase producing fungal strain Amesia atrobrunnea was screened from rotted wood samples and agro waste using ABTS method (Thakkar et al. 2020).
CONCLUSION
In the present study, a fungal stain Alternaria alternata was screened and investigated. The results showed that the fungal isolate Alternaria sp. MK855476.1 exhibited an ability to oxidize ABTS. Ahalo of dark green and purple was formed around the fungal colonies (positive for ABTS oxidation), indicating the presence of ligninolytic enzymes laccase. Due to their broad substrate rang and a large number industrial and biotechnological application of these multicopper enzymes has led to a terrible increase in the demand for this biocatalyst. Optimizing the condition for increasing laccase production and decolourization of dyes by Alternaria alternata will be further investigated in the future.
ACKNOWLEDGEMENTS
This study was financially supported by Shri Ramswaroop Memorial University Lucknow-Deva Road, Barabanki, Uttar Pradesh, India through University Research Fellow (URF).
Conflict of Interest: Authors declare no conflicts of interests to disclose.
REFERENCES
Abeer, A., E.l Aty., A., Aliaa, R., et al. (2015). Screening of fungal isolates for laccase enzyme production from marine sources. Res j pharm biol chem Sci., 6(1), 221-228.
Antecka, K., Zdarta, J., Siwińska-Stefańska, K., et al. (2018). Synergistic degradation of dye wastewaters using binary or ternary oxide systems with immobilized laccase. Catalys., 8(9), 402-420. https://doi.org/10.3390/ catal8090402
Das, A., Bhattacharya, S., Panchanan, G., et al. (2016). Production, characterization and Congo red dye decolourizing efficiency of a laccase from Pleurotus ostreatus MTCC 142 cultivated on co-substrates of paddy straw and corn husk. J Genet Eng Biotechnol, 14(2), 281-288. https://doi.org/10.1016/j.jgeb.2016.09.007
Deepa, T., Gangwane, A., Sayyed, R.Z. et al. (2020). Optimization and scale-up of laccase production by Bacillus sp. BAB-4151 isolated from the waste of the soap industry. J. Environ. Sustain. 3, 471–479. https://doi.org/10.1007/s42398-020-00135-9
Delgado-Baquerizo, M., Powell, J.R., Hamonts, K., et al. (2017). Circular linkages between soil biodiversity, fertility and plant productivity are limited to topsoil at the continental scale. New Phytol., 215, 1186-1196. https://doi.org/10.1111/nph.14634
Devasia, S., and Nair, A.J. (2016). Screening of potent laccase producing organisms based on the oxidation pattern of different phenolic substrates. Int. J Curr Microbiol App Sci., 5(5), 127-137. http://dx.doi.org/10.20546/ijcmas.2016.505.014
Dwivedi, UN., Singh, P., Pandey, VP., et al. (2011). Structure–function relationship among bacterial, fungal and plant laccases. J Mole Cat B Enz., 68(2), 117–128. https://doi.org/10.1016/j.molcatb.2010.11.002
Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evoln., 39(4), 783-791. doi: 10.1111/j.1558-5646.1985.tb00420.x.
Gassara, F., Brar, SK., Tyagi, RD., et al. (2010). Screening of agro-industrial wastes to produce lignolytic enzymes by Phanerochaete chrysosporium. Biochem. Eng. J., 49(3), 388-394. doi: 10.1016/j.bej.2010.01.015
Gedikli, S., Aytar, P., Unal, A., et al. (2010). Enhancement with inducers of lacasse production by some strains and application of enzyme to dechlorination of 2,4,5-trichlorophenol. Electron J Biotechnol.,13(6). https://doi.org/10.2225/vol13-issue6-fulltext-9
Glaeser, JA., and Lindner, DL. (2010). Use of fungal biosystematics and molecular genetics in detection and identification of wood-decay fungi for improved forest management. Forest Pathology, 41, 341-348. https://doi.org/10.1111/j.1439-0329.2010.00681.x
Hannula, SE., Morriën, E., Hollander, MD., et al. (2017). Shifts in rhizosphere fungal community during secondary succession following abandonment from agriculture. ISME J., 11(10), 2294-2304. doi: 10.1038/ismej.2017.90
Irfan, M., Mehmood, S., Irshad, M., et al. (2018). Optimized production, purification and molecular characterization of fungal laccase through Alternaria alternata, Turkish, J. Biochem., 43(6), 613-22. https://doi.org/10.1515/tjb-2017-0239
Kanaujia, A., Kumar, A., Kushwaha, S., et al. (2014). Status of Faunal Biodiversity and Threats to Wetlands of Barabanki District, Uttar Pradesh, India. Lif Sci., 2(4), 281-288.
Kimura, M., (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol Evoln., 16(2), 111-120. DOI: 10.1007/BF01731581
Margot, J., Maillard, J., Rossi, L., et al. (2013). Influence of treatment conditions on the oxidation of micropollutants by Trametes versicolor laccase. N Biotechnol., 30(6), 803–813. https://doi.org/10.1016/j.nbt.2013.06.004
Phatake, YB., Siddiqui, RA., Peshwe, SA., et al. (2015). Studies on degradation of synthetic dyes by using laccase producing Aspergillus nidulans isolated from textile effluent. I J life sci., 4(2), 67-78.
Pointing, SB., Jones, E.B.G., and Vrijimoed, L.L.B. (2000). Optimization of laccase production by Pycnoporus sanguieneus in submerged liquid culture. Mycologia., 92(1), 139-144. https://doi.org/10.1080/00275514.2000.12061138
Rao, A., Ramakrishna, N., Arunachalam, S., et al. (2019). Isolation, screening and optimization of laccase-producing endophytic fungi from Euphorbia milii. Arabian sci and Eng., 44(1), 51-64. https://doi.org/10.1007/s13369-018-3431-8
Saitou, N., and Nei, M., (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mole Biol and Evoln., 4(4), 406-425. doi: 10.1093/oxfordjournals.molbev.a040454
Senthivelan, T., Kanagaraj, J., Panda, R.C., et al. (2019). Screening and production of a potential extracellular fungal laccase from Penicillium chrysogenum: Media optimization by response surface methodology (RSM) and central composite rotatable design (CCRD). Biotecnol Rep., 23. https://doi.org/10.1016/j.btre.2019.e00344
Sharma, A., Aggarwal, NK., and Yadav, A., (2016). First report of lignin peroxidase production from Alternaria alternata ANF238 isolated from rotten wood sample. J Bioeng and Biosci., 4(5), 76-87. DOI: 10.13189/bb.2016.040502
Shleev, SV., Morozova, OV., Nikitina, OV., et al. (2004). Comparison of physico-chemical characteristics of four laccases from different basidiomycetes. Biochimie., 86(9-10), 693-703. https://doi.org/10.1016/j.biochi.2004.08.005
Tamura, K., Peterson, D., Peterson, N., et al. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mole Biol and Evoln., 28(10), 2731-2739. DOI: 10.1093/molbev/msr121
Tapwal, A., Varghese, S., Kumar, U., et al. (2014). Production of Laccase by Alternaria alternata and Lasiodiplodia theobromae. Eur. J. Exp. Biol., 4(4), 196-201.
Teerapatsakul, C., and Chitradon, L. (2016). Physiological regulation of an alkaline-resistant laccase produced by Perenniporia tephropora and efficiency in biotreatment of pulp mill effluent. Microbiol., 44(4), 260-268. https://doi.org/10.5941/MYCO.2016.44.4.260
Thakkar, A.T., Pandya, D.C. and Bhatt, S.A., (2020). Optimization of Laccase Enzyme Production by Amesia atrobrunnea A2: A First Report. Biosciences Biotechnology Research Asia, 17(1), 65-72.
Vantamuri, A.B., and Kaliwal, B.B., (2015). Isolation, screening and identification of laccase producing fungi. Int J Pharm Bio Sci., 6(3), 242-250.
Vera, M., Nyanhongo, GS., Pellis, A., et al. (2019). Immobilization of Myceliophthora thermophila laccase on poly (glycidyl methacrylate) microspheres enhances the degradation of azinphos-methyl. J Appl Polym Sci., 136(16), 1-10. 47417. DOI: 10.1002/app.47417
Viswanath, B., Rajesh, B., Janardhan, A., et al. (2014). Fungal laccases and their applications in bioremediation, Enzyme Res., 2014. 1-21. http://dx.doi.org/10.1155/2014/163242
Wang, Q., Qian, Y., Ma, Y., et al. (2018). A preliminary study on the newly isolated high laccase-producing fungi: screening, strain characteristics and induction of laccase production. Open Life Sci., 13(1), 463-469. https://doi.org/10.1515/biol-2018-0055
Xu, X., Xu, Z., Shi, S. et al. (2017). Lignocellulose degradation patterns, structural changes and enzyme secretion by Inonotus obliquus on straw biomass under submerged fermentation. Bioresour Technol., 241, 415-423. https://doi.org/10.1016/j.biortech.2017.05.08
Yadav, M., Bista, G., Maharjan, R., et al. (2019). Secretory laccase from Pestalotiopsis Species CDBT-F-G1 fungal strain isolated from high altitude, optimization of its production and characterization. Appl Sci., 9(2), 340. https://doi.org/10.3390/app9020340
Zdarta, J., Antecka, K., Frankowski, R., et al. (2018). The effect of operational parameters on the biodegradation of bisphenols by Trametes versicolor laccase immobilized on Hippospongia communis spongin scaffolds. Sci Total Environ., 615, 784-795. https://doi.org/10.1016/j.scitotenv. 2017.09.213
Zeng, S., Qin, X., and Xia, L. (2017). Degradation of the herbicide isoproturon by laccase mediator systems. J Biochem Eng., 119, 92-100. https://doi. org/10.1016/j.bej.2016.12.016


