1Department of Medical Laboratories, Riyadh Security Forces Hospital, Ministry of Interior, Riyadh, Saudi Arabia
2Department of Medical Microbiology and Parasitological, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia
3Clinical and Molecular Microbiology Laboratory King Abdulaziz University Hospital, Jeddah, Saudi Arabia
4Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
5Department of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia
6Department of Infectious Disease, King Abdulaziz University, Jeddah, Saudi Arabia
7Department of Infection Control and Environmental Health, King Abdulaziz University Hospital, Jeddah, Saudi Arabia
Corresponding author email: dr.dalia2007@hotmail.com
Article Publishing History
Received: 10/12/2020
Accepted After Revision: 25/03/2021
Bloodstream infection (BSI) is one of the primary causes of morbidity and mortality worldwide. The management of nosocomial BSI is challenging. BSI may be associated with Multidrug-resistant Gram-Negative Bacteria (MDR-GNB), which are difficult to treat with conventional and available antimicrobial drugs. Globally, the increased prevalence of MDR -GNB has led to a significant change in the spectrum of microorganisms isolated from patients with BSI. The aim of this study to investigate the prevalence , epidemiological aspects and Microbiological pattern of BSI caused by MDR-GNB at King Abdulaziz University Hospital in Jeddah, Saudi Arabia, to facilitate the development of Multidrug-Resistant Organisms (MDROs) Prevention and Control policy and to support proper selection of antimicrobial treatment and management of MDR-GNB infection .
Method: a retrospective analysis conducted in patients with GNB BSI, which included all hospital departments, using the data from the Clinical and Molecular Microbiology Laboratory database. All positive blood culture results from June 2017 to June 2020 were reviewed. Result: a total of 302 patients with positive blood culture were identified. The major risk factors for acquiring BSI were immunocompromised conditions, such as cancer (25%) and kidney disease (24.5 %,). The emergency room was the department with the most isolated cases (39.4%). Escherichia coli (43%) was the principal Gram Negative Bacilli responsible for BSI, and Acinetobacter baumannii was the most extensively drug-resistant GNB (84%). In conclusion, this study illustrates the importance and value of continuous surveillance of MDROs. Clinical microbiology laboratories should monitor MDR , XDR and Pan drug-resistance (PDR) bacterial strains to reduce the incidence of antimicrobial resistance and to help in the formulation of effective antimicrobial stewardship programmes in healthcare facilities.
Bloodstream Infection, Blood Culture, Multidrug-Resistant Gram-Negative Bacteria , Antimicrobial Resistance.
Aldossari R.O, Fatani A.J, Attallah D.M, Alhazmi W.A, Kaki R. Prevalence and Microbiological Pattern of Blood Stream Infection Caused by Multi Drug Resistance Gram Negative Bacteria in Western Saudi Arabia. Biosc.Biotech.Res.Comm. 2021;14(1).
Aldossari R.O, Fatani A. J, Attallah D. M, Alhazmi W. A, Kaki R. Prevalence and Microbiological Pattern of Blood Stream Infection Caused by Multi Drug Resistance Gram Negative Bacteria in Western Saudi Arabia. Biosc.Biotech.Res.Comm. 2021;14(1). Biosc.Biotech.Res.Comm. 2021;14(1). Available from: <a href=”https://bit.ly/3rkOiRa“>https://bit.ly/3rkOiRa</a>
Copyright © Aldossari et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Blood stream infection (BSI) and bacterial sepsis are public health threats. Recently World Health Organization listed BSI as a global health priority (Leal et al., 2019). The management and treatment of BSI have become challenging during the last decade due to the emergence of Multidrug-resistant organisms (MDROs) that are difficult to treat using conventional antimicrobial drugs (Gudiol et al., 2011). The increased prevalence of Multidrug Resistance Gram Negative Bacteria (MDR-GNB) has led to a significant change in the spectrum of microorganisms isolated from patients with BSI (Breijyeh et al., 2020). Understanding the definition and the mechanisms of antibiotic resistance and how these mechanisms can evolve and spread is essential for surveillance and tracking the spread of drug resistance Bacteria (Iredell et al., 2016).
MDROs is defined as non-susceptible or resistance of a microorganism to the antimicrobial agents in spite of previously susceptible to it (Tanwar et al., 2014). Basak et al. (2016) defined extensively drug-resistant (XDR) as bacteria non-susceptible to at least one drug in all but two or fewer antimicrobial categories (i.e., bacterial isolates sensitive to only one or two antimicrobial types); and pan drug-resistance (PDR) as no susceptibility to all agents in all antimicrobial categories. Infections due to MDR-GNB are an increasing threat to human health and are associated with excessive morbidity, mortality, and healthcare costs (Morris and Cerceo, 2020). It has become more challenging to control the spread of MDROs due to their growing antibiotic resistance (IDSA, 2011). A rising attention about the clinical and economic impact of MDROs has led to a major focus on antibiotic stewardship to reduce inappropriate antimicrobial prescribing (Thatrimontrichai, et al., 2020).
MATERIAL AND METHODS
2.1 Study Design: A retrospective study for all patients with Gram Negative Bacteria (GNB) BSI was conducted at King Abdulaziz University Hospital (KAUH) in Jeddah, Saudi Arabia. All departments of KAUH (ER, ICU/CCU, MMW, MICU, FMW, NICU,PW & SICU) were included in our study. The sample population included all age groups. All positive blood culture results from June 2017 to June 2020 were reviewed using study data obtained from the Clinical and Molecular Microbiology Laboratory (CMML) database.
We only considered blood cultures and did not include other types of microbiology culture. If a patient had multiple admissions for GNB, they were included in the study as different episodes. However, if a patient developed recurrence of BSI during the same admission, it was considered as a single-patient episode of BSI. There were no other exclusion criteria.
Patients’ electronic medical records were reviewed. Data collection included the following clinical variables: (a) age and gender (b) comorbid conditions; (c) use of antibiotics in the last 30 days; (d) source of infection; (e) use of a central venous catheter ≥48 h before the onset of GNB (f) antimicrobial resistance patterns in GNB blood culture isolates; and (g) mortality within 30 days. Ethical approval for all patients was obtained from the KAUH Research Ethics Committee (reference no.: 543-20 Oct.29.2020). The requirement of patient consent was waived due to the retrospective nature of the study.
2.2 Study Definitions
The following definitions were used: MDROs were defined according to the US Centres for Disease Control and Prevention definitions. MDR-GNB were defined as ESBL-producing Enterobacteriaceae and any GNB (e.g., Acinetobacter spp., Enterobacteriaceae, and Pseudomonas spp.) resistant to three or more of the following drug classes: piperacillin/tazobactam, Cephalosporins (Cefazolin, Ceftriaxone, Ceftazidime, and Cefepime), Carbapenems (Imipenem), Monobactams (Aztreonam), Aminoglycosides (Gentamicin, Tobramycin, and Amikacin), and Fluoroquinolones (Ciprofloxacin and Levofloxacin). Recurrence of BSI was defined as a positive blood culture with same GNB after ≥1 negative blood culture and after an interval of ≥7 days. Mortality was defined as death by any cause within 30 days of the onset of BSI.
2.3 Identification and characterisation of the bacterial isolates: Blood culture bottles were incubated at CMML using the BacT/Alert VIRTUO Microbial Detection System (bioMérieux, Durham, NC, USA), which is fully automated and yields real-time results. The blood culture bottles were incubated until a signal-positive alarm was sounded or for a maximum of 5 days. Samples from the positive blood culture bottles were processed using Gram staining, the results were entered in the system and the department was verbally informed.
Then, following the CMML’s blood culture manual, all positive blood culture bottles were sub-cultured on 5% sheep blood agar, chocolate agar and MacConkey agar (Saudi Prepared Media Laboratories). The MacConkey agar plates were incubated at 35–37°C for 18–24 h in an ordinary incubator (Forma Scientific Incubator, Germany). The blood agar and chocolate agar plates were incubated at 35–37°C in 5–10% CO2 (Sanyo CO2 Incubator, Japan).
Antibiotic sensitivity was assessed using a manual technique (the disc diffusion method). Mueller-Hinton plates (Saudi Prepared Media Laboratories, Riyadh, Saudi Arabia) were inoculated with blood samples taken directly from the positive blood culture bottles. The plates were incubated at 35–37°C for 18–24 hours in an ordinary incubator (Forma Scientific Incubator). Antibiotic discs were selected according to the guidelines provided by the Clinical and Laboratory Standard Institute (CLSI).
After 24 h of incubation, gram-negative bacilli colonies were identified using a VITEK 2 system (bioMérieux, Marcy-L’Étoile, France) according to the manufacturer’s instructions. This automated system uses a turbid metric method with VITEK 2 GN ID (BioMérieux), namely Gram-negative identification cards including members of the family Enterobacteriaceae as well as non-enteric bacilli. The suspension was prepared from a pure sub-culture plate by mixing the colony with 3.0 mL of 0.45% sterile saline, which was aseptically added to the plastic test tube. Density was measured by a VITEK 2 DensiCheck System (bioMérieux), and results equivalent to 0.5–0.63 of McFarland standards were used.
The suspension tube was placed in a cassette and followed by an empty tube. The VITEK 2 ID Card was inserted in the suspension tube. Less than 30 min elapsed between the preparation of the suspension and the card filling. The cassettes were then loaded into the VITEK 2 system. When the process was completed, on board software and automation moved the cards to the discard area after analysing the data. Finally, the results were collected from the VITEK 2 system after 10–18 h. When the sample cycle was finished, the used cards were discarded in a biohazard bag.
2.4 Antimicrobial susceptibility testing: The VITEK 2 system was used for antibiotic susceptibility testing. AST-GN susceptibility cards (panels N91 and N92) were used according to the manufacturer’s instructions. The VITEK 2 system controlled the cards automatically, including their filling, sealing, and transfer to the incubator (35°C). Each AST-gram-negative susceptibility card was placed next to a VITEK 2 card in an empty tube.
The results were collected from the VITEK 2 system after 10–18 h. When the sample cycle was finished, the VITEK 2 cassette and tube were discarded in a biohazard bag. The results from the VITEK 2 system were compared to the Gram-negative bacteria identification databank. CMML’s antibiotic susceptibility reporting criteria for interpreting resistance, sensitivity and intermediate resistance were based on the updated guidelines of the CLSI. A renewal of that guideline is made with the issuance of each new annual edition by CLSI.
2.5 Data analysis: All data were analysed using SPSS version 22 statistical software (IBM Corp., Armonk, NY, USA). Numerical data were reported as mean ± standard deviation, and categorical data were reported using frequencies and percentages. Chi-square test was used to assess the significance of associations between the study variables and the pathogen types. P-values < 0.05 were considered significant.
RESULTS
3.1 Demographic and clinical characteristics: A total of (302) patients were included in the analysis. As shown in Table (1), the numbers of males and females were similar. The majority of the patients (n=170, 56%) were aged >50-years. Most of the BSI cases were obtained from the emergency room (ER). Within the sample population, the groups with highest number of BSI were patients diagnosed with immunocompromised conditions such as cancer (25%), or kidney disease (24.5%). Most of the BSIs (65%) had an exovascular infection route as secondary infections. The overall mortality rate of the study population was considerably high (n=160, 53%).
Table 1. Epidemiological and clinical characteristics of (302) patients diagnosed as BSI associated with MDR-GNB strain in a period from June 2017 to June 2020 in KAUH.
| Demographic Characteristics | No (%)
n=302 |
| Age (years) | 46.9 ± 28.2 (54) |
| Age groups (years)
0–2 2–18 18–50 >50 |
48 (15.9%) 19 (6.3%) 65 (21.5%) 170 (56.3%) |
| Sex
Male Female |
153 (50.7%) 149 (49.3%) |
| KAUH Department
ER ICU/CCU MMW MICU FMW NICU PW SICU |
119 (39.4%) 22 (7.3%) 20 (6.6%) 51 (16.9%) 15 (5%) 12 (4%) 25 (8.3%) 12 (4%) |
| Clinical Characteristics | No (%)
n=302 |
| (immunocompromised patients)
Cancer Heart disease Pulmonary disease Kidney disease Sepsis and meningitis Liver diseases (cirrhosis) Diabetes mellitus
|
76 (25%) 38 (12.6%) 40 (13.2%) 74 (24.5%) 10 (3.3%) 7 (2.3%) 14 (4.6%) |
| Infection Route
Exovascular Endovascular Not determined |
197 (65.2%) 100 (33.1%) 5 (1.7%) |
| Number of Deaths | 160 (53%) |
All numerical data are presented as mean ± standard deviation (median). All categorical data are presented in n (%).
Abbreviations: CCU, coronary care unit; ER, emergency room; FMW, female medical ward; ICU, intensive care unit; MICU, medical intensive care unit; MMW, male medical ward; NICU, neonatal intensive care unit; SICU, surgical intensive care unit; PW,Peadiatric ward.
3.2 Microbial spectrum and susceptibility patterns of pathogens causing bloodstream infections
From figure (1) E.coli was the most prevalence GNB organisms causing BSI (n=130, 43%) & the second most common organisms causing BSI was K. pneumoniae (n=94, 31%) . Nearly 97% of the E. coli were ESBL producers (Table 2), and 77% were resistant to Ciprofloxacin (Table 3). Moreover, 90% of the K. pneumoniae were ESBL producers, while only 5% were CRE (Table 2).
aeruginosa occurred less frequently than GNB BSIs due to the other major organisms (about 9%, p<0.001). The prevalence of MDR was highest among P. aeruginosa (66%, p<0.001) (Table 2). Furthermore, the susceptibility pattern of P. aeruginosa showed a higher prevalence of Imipenem resistance (63%) (Table 3), resulting in 33% of the isolates being reported as carbapenem-resistant P. aeruginosa. A further 51 cases (17%) were caused by A. baumannii (Figure 1), of which 84% of the isolates were extensively drug resistant (XDR) (p<0.001) (Table 2).
Table 2. Distribution of MDR-GNB causing BSI. Data collect during a period from June 2017 to June 2020 in KAUH.
| Pathogen | Total | Percentage | ESBL | MDR | XDR | CRP | CRE | P-value |
| E. coli | 130 | 43.0% | 126 (96.9%) | 1 (0.8%) | 0 | 0 | 3 (2.3%) | <0.001 |
| K. pneumoniae | 94 | 31.1% | 85 (90.4%) | 4 (4.3%) | 0 | 0 | 5 (5.3%) | |
| P. aeruginosa | 27 | 8.9% | 0 | 18 (66%) | 0 | 9 (33%) | 0 | |
| A. baumannii | 51 | 16.9% | 0 | 6 (12%) | 43 (84%) | 0 | 0 |
The P-value was calculated using the chi-square test. Values <0.05 are statistically significant.
Abbreviations: A. baumannii, Acinetobacter baumannii; CRE, carbapenem-resistant Enterobacteriaceae; CRP, carbapenem-resistant P. aeruginosa; E. coli, Escherichia coli; ESBL, extended-spectrum beta-lactamase producers; K. pneumoniae, Klebsiella pneumoniae; MDR, multidrug-resistant; P. Aeruginosa, Pseudomonas aeruginosa; XDR, extensively drug-resistance
Table 3. Susceptibility patterns of multidrug-resistant Gram-Negative Bacteria (GNB) causing BSI .
| GNB | TZP | CAZ | CRO | IMP | MEM | CIP | GM | AK | CO |
| E. coli n=130 (%) |
129 (99) | 130 (100) | 130 (100) | 3 (2.3) | 3 (2.3) | 100 (77) | 44 (34) | 1 (0.8) | 0 |
| K. pneumoniae n=94 (%) |
94 (100) | 94 (100) | 94 (100) | 3 (3.2) | 3 (3.2) | 60 (64) | 38 (40) | 10 (11) | 0 |
| P. aeruginosa n=27 (%) |
21 (78) | 19 (70.4) | 0 | 17 (63) | 17 (63) | 15 (56) | 6 (22.2) | 5 (18.5) | 0 |
| A. baumannii n=51 (%) |
0 | 50 (98) | 0 | 50 (98) | 50 (98) | 50 (98) | 40 (78) | 39 (76) | 2 (4) |
Abbreviations: A. baumannii, Acinetobacter baumannii; AK, Amikacin; CAZ, Ceftazidime; CIP, Ciprofloxacin; CO, Colistin;CRO, Ceftriaxone; E. coli, Escherichia coli; IMP. Imipenem; GM, Gentamicin; GNB ,Gram Negative Bacteria ; MEM, Meropenem; K. pneumoniae, Klebsiella pneumoniae; P. aeruginosa, Pseudomonas aeruginosa; TZP,Pipracillin _ tazobactam.
Figure 1: The microbial spectrum of multidrug-resistant gram-negative bacteria causing bloodstream infections during a period from June 2017 to June 2020 in KAUH.
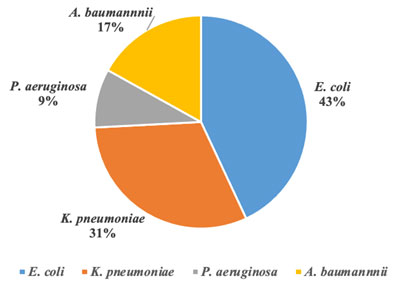
Abbreviations: A. baumannii, Acinetobacter baumannii; E. coli, Escherichia coli; K. pneumoniae, Klebsiella pneumoniae; &P. aeruginosa, Pseudomonas aeruginosa.
3.3 Major risk factors for BSI :The three most common risk factors leading to BSI were an impaired immune system, an underlying chronic disease, and older age (Table 4). In addition, 26% of the patients had indwelling devices.
Table 4. Major risk factors for BSI among patients in KAUH
| Pathogen | Immunocompromised Patients | Underlying Chronic Diseases | Aged >65 Years | Indwelling Devices |
| E. coli n=130 (%) |
100 (77%) | 110 (85%) | 79 (60.8%) | 42 (32.3%) |
| K. pneumoniae n= 94 (%) |
62 (66%) | 78 (83%) | 47 (50%) | 21 (22.3%) |
| P. aeruginosa n=27 (%) |
21 (78%) | 24 (89%) | 12 (44.4%) | 6 (22.2%) |
| A. baumannii n=51 (%) |
38 (74.5%) | 41 (80.4%) | 32 (62.7%) | 14 (27.5%) |
DISCUSSION
BSI are a significant cause of morbidity and mortality worldwide. Over the last decades, there has been a significant increase in the number of pathogen isolated from BSI cases that are resistant to antimicrobial drugs (Leal et al., 2019). Worldwide, numerous MDROs are the leading causes of nosocomial infections (Exner et al., 2017). On other hand, the incidence of community-acquired MDR-GNB infection has also been increasing (Tseng et al., 2017). In our study, the hospital department with the highest number of MDR-GNB infections was the Emergency Room whereas; all the patients arrived to this department were from different segments of the community.
Recognising the risk factors in the development of MDR-GNB in BSI could significantly influence patient management. (Bassetti et al. 2017) reported that the factors that have contributed to the spread of MDR-GNB include the overuse of existing antimicrobial drugs, which has promoted the development of adaptive resistance mechanisms by bacteria. BSI is a life-threatening condition, especially for vulnerable individuals, such as those who are immunocompromised, older adults and individuals with underlying diseases (Exner et al., 2017).
Similarly, our study found that underlying chronic diseases and impaired immune systems were major predisposing factors in the development of MDR-GNB and that the groups with the highest frequency of BSIs were immunocompromised patient cases, such as cancer (25%) and kidney diseases (24.5%). MDR-GNB is common among residents in long-term care facilities, particularly those residents with indwelling devices, and these facilities are an important source of such strains among patients admitted to healthcare facilities (Kaye and Pogue, 2015).
In our study, we found that over one-quarter of the patients with MDR-GNB infections had a history of an indwelling device used.According to Kuntaman et al. (2018), most patients with MDR-GNB are seriously ill and have a poor prognosis with a high mortality rate. As it shown as evident in the results of our study, the mortality was increased in BSI associated with MDR-GNB (53 %).
In MDR-GNB counting, the non-fermenter GNB have a lower frequency of isolation than Enterobacteriaceae such as Escherichia coli (E.coli) and Klebsiella pneumonia (K. pneumonia), while the primary non-fermenter GNB that cause human infections are Acinetobacter baumannii (A. baumannii) and Pseudomonas aeruginosa (P. aeruginosa) (Oliveira and Reygaert, 2019). The prevalence of MDR E. coli strains is rising worldwide (Allocati et al., 2013) .The most common MDR-GNB in our study were E. coli, followed by K. pneumoniae, A. baumannii, and P. aeruginosa.
Ruppé et al. (2015) determined that Enterobacteriaceae, were the most important MDR-GNB and that dramatic increase drug-resistance trend in most of the anti-gram-negative agents (β-lactams, Fluoroquinolones, and Aminoglycosides) was the most important resistance issue.
Rawat and Nair (2010) determined that extended-spectrum β-lactamases (ESBLs) were a mechanism by which the GNB developed antibiotic resistance in the face of introduction of new antimicrobial agents. ESBLs efficiently hydrolyse extended-spectrum β-lactams, such as Cefotaxime, Ceftriaxone, Ceftazidime, and Aztreonam.
E. coli and K. pneumoniae are the most prevalent members of the Enterobacteriaceae group and are responsible for widespread ESBL production such as: SHV-1, TEM-1, and TEM-2 (Al-Otaibi et al., 2016). In our study too, the E. coli ESBL producers were the predominant isolates among the GNB-causing BSI
Carbapenems, such as Imipenem and Meropenem, which are classes of β-lactam, are the most effective treatments for infections caused by ESBL-producing bacteria (Breijyeh et al., 2020).Carbapenem-resistant Enterobacteriaceae (CRE) or carbapenemase-producing Enterobacteriaceae, Acinetobacter baumannii (CRAB), and Pseudomonas aeruginosa (CRPsA) are earnest cause of nosocomial infections (Tomczyk et al., 2019).
baumanniiand P. aeruginosa are increasingly acquiring carbapenem resistance which, given their intrinsic antibiotic resistance, can cause difficult-to-treat infections (Gniadek et al., 2016). Zhang et al. (2016) reported that P. aeruginosa can cause severe infections, such as BSI, with a high prevalence of Carbapenem resistance. In our study, the resistance to Imipenem and Meropenems was low for E. coli and K. pneumonia but high for A. baumannii and P. aeruginosa .
Due to a variable resistance mechanism, such as altering the target position (penicillin-binding proteins), the development of β-lactamase, the narrowing of membrane permeability, and efflux pump, A. baumannii MDR infections are difficult to treat, owing to the extremely limited armamentarium (Lee et al., 2017). This is evident from the results of our study on this type of GNB, wherein most of the A. baumannii isolates were of XDR strains.
Limitations of our study include the following: (1) it was a single-centre study; (2) it was based on the retrospective analysis of clinical data and (3) the time to source control, which can impact the mortality rate, was not assessed.
CONCLUSION
This study found a rise in the prevalence of GNB-MDR, highlighting the importance of continuous surveillance for the proper care of patients with relevant risk factors and choosing the best empirical treatment. To manage and cure hospitalised patients appropriately, it is vital to identify the bacteria responsible for the infection and their antimicrobial susceptibility profiles.
We recommend that all clinical microbiology laboratories implement early detection and close monitoring of MDR, XDR and PDR bacterial strains to reduce the problem of antimicrobial resistance – now a global problem – and help in the formulation of effective antimicrobial stewardship programmes in healthcare facilitie.
REFERENCES
Allocati, N., Masulli, M., Alexeyev, M. F., & Di Ilio, C. (2013). Escherichia coli in Europe: an overview. International journal of environmental research and public health, 10(12), 6235–6254.
Al-Otaibi, F. E., Bukhari, E. E., Badr, M., & Alrabiaa, A. A. (2016). Prevalence and risk factors of Gram-negative bacilli causing blood stream infection in patients with malignancy. Saudi medical journal, 37(9), 979–984.
Basak S, Singh P, Rajurkar M. Multidrug Resistant and Extensively Drug Resistant Bacteria: A Study. J Pathog. 2016;2016:4065603.
Bassetti, M., Poulakou, G., Ruppe, E., Bouza, E., Van Hal, S. J., & Brink, A. (2017). Antimicrobial resistance in the next 30 years, humankind, bugs and drugs: a visionary approach. Intensive care medicine, 43(10), 1464–1475.
Breijyeh, Z., Jubeh, B., & Karaman, R. (2020). Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules (Basel, Switzerland), 25(6), 1340.
Exner, M., Bhattacharya, S., Christiansen, B., Gebel, J., Goroncy-Bermes, P., Hartemann, P., Heeg, P., Ilschner, C., Kramer, A., Larson, E., Merkens, W., Mielke, M., Oltmanns, P., Ross, B., Rotter, M., Schmithausen, R. M., Sonntag, H. G., & Trautmann, M. (2017). Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria?. GMS hygiene and infection control, 12, Doc05.
Gniadek, T. J., Carroll, K. C., & Simner, P. J. (2016). Carbapenem-Resistant Non-Glucose-Fermenting Gram-Negative Bacilli: the Missing Piece to the Puzzle. Journal of clinical microbiology, 54(7), 1700–1710.
Gudiol, C., Tubau, F., Calatayud, L., Garcia-Vidal, C., Cisnal, M., Sánchez-Ortega, I., Duarte, R., Calvo, M., & Carratalà, J. (2011). Bacteraemia due to multidrug-resistant Gram-negative bacilli in cancer patients: risk factors, antibiotic therapy and outcomes. The Journal of antimicrobial chemotherapy, 66(3), 657–663.
Infectious Diseases Society of America (IDSA), Spellberg, B., Blaser, M., Guidos, R. J., Boucher, H. W., Bradley, J. S., Eisenstein, B. I., Gerding, D., Lynfield, R., Reller, L. B., Rex, J., Schwartz, D., Septimus, E., Tenover, F. C., & Gilbert, D. N. (2011). Combating antimicrobial resistance: policy recommendations to save lives. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 52 Suppl 5(Suppl 5), S397–S428.
Iredell, J., Brown, J., & Tagg, K. (2016). Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. Bmj, 352.
Leal, H.F., Azevedo, J., Silva, G.E.O., Amorim, A.M.L., de Roma, L.R.C., Arraes, A.C.P., and Reis, J.N. (2019). ‘Bloodstream infections caused by multidrug-resistant gram-negative bacteria: Epidemiological, clinical and microbiological features’, BMC Infectious Diseases, 19(1), pp. 1-11.
Kaye, K. S., & Pogue, J. M. (2015). Infections Caused by Resistant Gram-Negative Bacteria: Epidemiology and Management. Pharmacotherapy, 35(10), 949–962.
Morris, S., & Cerceo, E. (2020). Trends, Epidemiology, and Management of Multi-Drug Resistant Gram-Negative Bacterial Infections in the Hospitalized Setting. Antibiotics (Basel, Switzerland), 9(4), 196.
Kuntaman, K., Shigemura, K., Osawa, K., Kitagawa, K., Sato, K., Yamada, N., Nishimoto, K., Yamamichi, F., Rahardjo, D., Hadi, U., Mertaniasih, N. M., Kinoshita, S., Fujisawa, M., & Shirakawa, T. (2018). Occurrence and characterization of carbapenem-resistant Gram-negative bacilli: A collaborative study of antibiotic-resistant bacteria between Indonesia and Japan. International journal of urology : official journal of the Japanese Urological Association, 25(11), 966–972.
Rawat D, Nair D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J Glob Infect Dis. 2010 Sep;2(3):263-74.
Ruppé, É., Woerther, P. L., & Barbier, F. (2015). Mechanisms of antimicrobial resistance in Gram-negative bacilli. Annals of intensive care, 5(1), 1-15.
Tanwar, J., Das, S., Fatima, Z., & Hameed, S. (2014). Multidrug resistance: an emerging crisis. Interdisciplinary perspectives on infectious diseases, 2014, 541340.
Tseng, W. P., Chen, Y. C., Yang, B. J., Chen, S. Y., Lin, J. J., Huang, Y. H., Fu, C. M., Chang, S. C., & Chen, S. Y. (2017). Predicting Multidrug-Resistant Gram-Negative Bacterial Colonization and Associated Infection on Hospital Admission. Infection control and hospital epidemiology, 38(10), 1216–1225.
Thatrimontrichai, A., & Apisarnthanarak, A. (2020). Active surveillance culture program in asymptomatic patients as a strategy to control multidrug-resistant gram-negative organisms: What should be considered?. Journal of the Formosan Medical Association = Taiwan yi zhi, 119(11), 1581–1585.
Tomczyk, S., Zanichelli, V., Grayson, M. L., Twyman, A., Abbas, M., Pires, D., Allegranzi, B., & Harbarth, S. (2019). Control of Carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in Healthcare Facilities: A Systematic Review and Reanalysis of Quasi-experimental Studies. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 68(5), 873–884.
Yunquera-Romero, L., Márquez-Gómez, I., Henares-López, A., Morales-Lara, M. J., Gallego Fernández, C., & Asensi-Díez, R. (2018). Adecuación de las prescripciones antimicrobianas realizadas en el área de urgencias de un hospital de tercer nivel [Appropriateness of antimicrobial prescriptions in the emergency department of a tertiary hospital]. Revista espanola de quimioterapia : publicacion oficial de la Sociedad Espanola de Quimioterapia, 31(3), 209–216.
Zhang, Y., Chen, X. L., Huang, A. W., Liu, S. L., Liu, W. J., Zhang, N., & Lu, X. Z. (2016). Mortality attributable to carbapenem-resistant Pseudomonas aeruginosa bacteremia: a meta-analysis of cohort studies. Emerging microbes & infections, 5(3), e27.


