1Manonmaniam Sundaranar University, Abishekapatti, Tirunelveli, India
2Sur University College, Sultanate of Oman
3B S Abdur Rahman Crescent Institute of Science and Technology, Chennai, India
Corresponding author email: nishajoseph226@gmail.com
Article Publishing History
Received: 14/10/2020
Accepted After Revision: 11/12/2020
The main purpose of this paper is to develop a novel deep feature retrieval approach for performing brain tumor identifying process from Magnetic Resonant images. Brain tumor is caused due to uncontrolled cell divisions. Tumor detection in the early phase is very important which is useful for diagnosis and treatment of tumor. First the input Magnetic Resonant brain image is denoised by using the Modified Decision Based Unsymmetric Trimmed Median Filter (MDBUTMF) and then the image contrast is improved with the Contrast Limited Adaptive Histogram Equalization (CLAHE). After pre-processed the input image the next step is to retrieve the features from the denoised and contrast enhanced image. To extract the features from the pre-processed image this project proposed one novel feature retrieval technique named Deep Efficient Reduced Local Derivative Pattern (DERLDP).
After extracting the deep features, the next step is to partition the brain tumor based on these extracted features. To do this process the supervised segmentation approach is employed. Among several supervised segmentation approaches this work uses deep machine learning approach named Convolution Neural Network (CNN). Finally, the extracted features are given as input to these machine learning approach to partition the brain tumor regions. To find out the performance of the proposed deep feature retrieval and deep machine learning approach, four performance metrics are employed namely, Dice Similarity Coefficient (DSC), Positive Predictive Value (PPV), Jaccard Index (JI) and Sensitivity (SEN). From the experimental results, it is shown that the novel DERLDP and CNN performs better than other existing approaches.
Brain Tumor, Clahe, Mdbutmf, Derldp and Cnn.
Joseph N, Murugan D, Thomas B. J, Ramya A. A Novel Efficient Deep Feature Extractor and Classifier Approach for Brain Tumor Segmentation in Magnetic Resonant Images. Biosc.Biotech.Res.Comm. 2020;13(4).
Joseph N, Murugan D, Thomas B. J, Ramya A. A Novel Efficient Deep Feature Extractor and Classifier Approach for Brain Tumor Segmentation in Magnetic Resonant Images. Biosc.Biotech.Res.Comm. 2020;13(4). Available from: <a href=”https://bit.ly/3m4aBYA”>https://bit.ly/3m4aBYA</a>
Copyright © Joseph et al., This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Brain tumor is a severe illness, where abnormal growths in the brain may interfere with the function of the brain. The National Fat Loss Foundation has announced that the number of persons in developing nations with brain tumors has increased by nearly 300 percent over the last three decades, (Logeswari, 2010; Lei 2016). Magnetic stereoscopic images are advanced medical image techniques that provide much data about the anatomy of soft tissue. Manual diagnosis of a brain tumor by a physician is a less-than-valid, long-term method (Menze 2014; Machhale 2015; Madabhushi 2016). The automatic detection of brain tumors using magnetic resonant images (MRIs) aspire to categorize usual and unusual MRIs based on the nonexistence or existence of tumors correspondingly. So, diagnosing problems can be considered a challenge for image classification. The widespread brain tumor has led to large MR data creation. Thus, the evolution of a self-healing brain tumor system that receives high accuracy of tumor detection and localization is essential. Full machine learning and deep learning play an important role in brain scanning, division, registration and classification of tumor tissue, (Pan 2015; Şentaş 2020).
In this section, we have propelled a few summaries of such existing studies and techniques. In Classification, the Fuzzy C-Means (FCM) segmentation is applied to separate the tumor and non-tumor region of brain. Also, wavelet feature is extracted by using multilevel Discrete Wavelet Transform (DWT). Finally, Deep Neural Network (DNN) is incorporated for brain tumor classification with high accuracy. This technique is compared with KNN, Linear Discriminant Analysis (LDA) and Sequential Minimal Optimization (SMO) classification methods. An accuracy rate of 96.97% in the analysis of DNN based brain tumor classification. But the complexity is very high and performance is very poor. New multi-fractal (MultiFD) feature retrieval and improved AdaBoost classification schemes are employed to identify and partition the brain tumor. The texture of brain tumor tissue is extracted by using MultiFD feature retrieval scheme. The improved AdaBoost classification methods are employed to find the given brain tissue is tumor or non-tumor tissue. Complexity is high. Local independent projection-based classification (LIPC) method is employed to classify the voxel of the brain (Şentaş 2020).
Also, path feature is extracted in this method. Hence no need to perform explicit regularization in LIPC. The accuracy is low. In MRI Images, a seeded tumor segmentation method with new Cellular Automata (CA) technique is presented, which is compared with graph cut based segmentation method. The seed selection and Volume of Interest (VOI) is calculated for efficient brain tumor segmentation. Also, tumor cut segmentation is incorporated into this work. The complexity is low. The accuracy is also low (El-Dahshan 2014; Ahmmed 2017). Researchers have suggested a strategy that accomplishes tumor stage by utilizing ANN. The Brain Tumor arrange is ordered utilizing the ANN classifier. The precision of the proposed strategy was expected around 97.44% (Ahmmed 2017). Researchers have contrived a novel strategy for brain tumor identification that envelops Histogram Normalization and selection of K-implies/ K means Segmentation schemes (Singh et al.2016). MRIs can be productively grouped the SVM in order to offer exact expectation and characterization.
SVM classifier allegedly gave the precision of 91.49%. As obviously apparent, the SVM approach offered higher precision. Researchers have received a scholarly grouping framework to sort normal and abnormal MR brain. In the classification step, diverse machine learning strategies like SVM, KNN and SVM-KNN have been embraced and a similar report among them is encouraged (Nandpuru 2016). Researchers have come up with an algorithm that is a mix of SVM and fuzzy c-implies, a hybrid scheme for recognition of brain tumor from MR scans (Parveen 2015). Researchers uses histogram, which computes the total quantity of specified pixel values distributed in a particular image (Sarma 2012). The algorithm was tested on 48 images where the overall accuracy rates for all images were around 95%. An advanced brain tumor segmentation was introduced, which was also called multimodal brain tumor segmentation scheme (LeCun 2010).
Hybrid feature selection with ensemble classification was applied for brain tumor diagnosis process. The GANNIGMAC, decision Tree, Bagging C based wrapper approach was employed to obtain the decision rules. The DeepSeg developed by Zeineldin et al. (2020) is a modular decoupling framework. It consists of two connected core parts based on an encoding and decoding relationship. The encoder part is a convolutional neural network (CNN) responsible for spatial information extraction. The resulting semantic map is inserted into the decoder part to get the full-resolution probability map. Automated segmentation using CNN has been proposed by Bhandari et al. (2020). CNNs work by using an input, convoluting this input with a filter (also termed a kernel) and giving an output. Wentao Wu et al. (2020) proposes a deep convolutional neural network fusion support vector machine algorithm (DCNN-F-SVM).
The proposed brain tumor segmentation model is mainly divided into three stages. In the first stage, a deep convolutional neural network is trained to learn the mapping from image space to tumor marker space. In the second stage, the predicted labels obtained from the deep convolutional neural network training are input into the integrated support vector machine classifier together with the test images. The advantages of the convolutional neural network are the fact that it provides optimal accuracy of segmentation. However, this is at the cost of computational load. To overcome this drawback, this paper introduces a novel feature extracting approach (Bhandari et al. 2020).
The main contributions of this paper are summarized as follows:
- CNN classifier was applied to partition the brain tumor region from the given input MR image. CNN classifier was trained by the brain MR image dataset BRATS datasets.
- For extracting features, a novel deep feature was employed named Modified Decision Based Unsymmetric Trimmed Median Filter (MDBUTMF)
- CNN classifier was compared with PNN and ANN classifiers.
- The proposed method provided good results for BRATS data set.
In the next section, we have proposed our methodology to identify and classify brain tumor from brain MR images that we have deduced by overcoming the found limitations on the subject.
MATERIAL AND METHODS
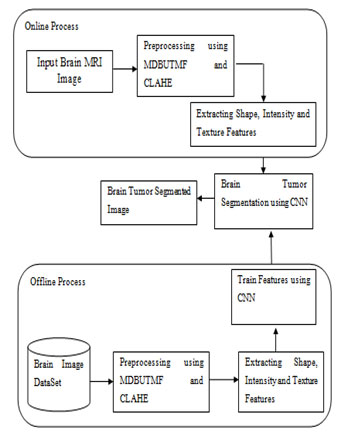
The overall architecture for the brain tumor segmentation based supervised learning has been shown in Fig. 1. First the input MR brain image is denoised by using MDBUTMF and then the image contrast is enhanced by using the CLAHE. After pre-processed the input image the next step is to extract the features from the denoised and contrast enhanced image. To extract the features from the pre-processed image this project proposed one novel feature retrieval technique named Deep Efficient Reduced Local Derivative Pattern (DERLDP). After extracting these deep features, the next step is to partition the brain tumor based on these extracted features. To do this process the supervised segmentation approach is employed. Among sever supervised segmentation approach this works uses deep machine learning approaches named CNN. Finally, the extracted features are given as input to these machine learning approaches to partition the brain tumor regions.
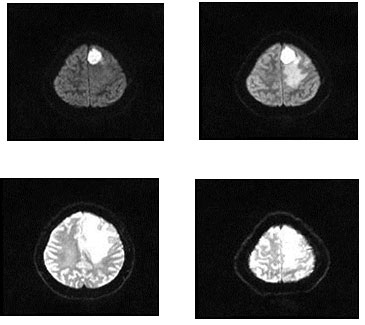
For the dataset used, the machine learning methods presented in this paper was evaluated on the BRATS 2015 databases. These datasets are downloaded from the site. A total of 4,800 images of the brain tumors were evaluated. The proposed technique is employed to the image of the tumor in the brain. During the training phase, a 4,000-millimeter brain tumor was employed, but each had 1000 pieces of brain tumors. It has five types of tumors, and each image has 200 images displayed. Experiments are carried out on a BRATS2015 FLAIR, T1, T1c and T2 training ground truth database of 4000 MR images of brain tumor and 800 testing ground truth databases. Fig. 2 depicts the sample brain tumor images from the BRATS 2015 database.
Figure 1: Overall Architecture of the Proposed Work
Figure 2: Experimental Images
For preprocessing, first, for the primary infrared image of the MR brain is enhanced by adaptive techniques. Analogue alignment (AHE) is a computer image processing technology employed to increase image levels. It differs from the simple histogram alignment, because many histogram manipulation methods, each corresponding to a particular part of the image, use them to distribute the light of the image. So, it is suitable for improving the local contrast and improving the edge definitions in each area of the image. However, AHE has a tendency to exaggerate noise in areas with the same image. The variant isotope variant called CLAHE, as opposed to the histogram, changes it by limiting the profit margin. Chip algorithms are shown in # 1 algorithm.
Algorithm1:
- Get al.l inputs: images, number of fields in rows and rows, histogram containers to use to create variable image function (dynamic range), limit delimation limit (normal 0 to 1)
- Prior to processing input: Set the exact limit of the clip from the normal value, if necessary, adjust the image before dividing it into the area.
- Process each area context (tile), and be able to produce image of grayscale: the single-zone retrieval of the image, creating the graph of the area using the number of bins, the histogram snap using the set of the set Create conversion (changes function) for this area.
- Correct image of gray level, in order to collect the final image of CLAHE: extracted from clusters of four adjacent features of the image processing map area by each tile-over-part artwork, download the application Pixel of Four Map Exercises to the Pixel and Interpolate Between Results to Get This Pixel Out! Repeat the whole image.
Next, for pre-processing, Modified Decision Based Unsymmetric Trimmed Median Filter (MDBUTMF) is employed on an enhanced MR brain images for de-noising. This filter processes corrupted images by identifying noise. The pixel process checks whether it is strong or not. That means that if the pixel runs between the maximum and minimum values of the gray level, it is considered to be in pixel, it is still unchanged. If the PPP process takes up the maximum green level, and the minimum is a puzzling pixel running by MDBUTMF.
For feature extraction, after denoising and contrast enhancement process, the next process is to retrieve the features from the preprocessed images. This work introduced one innovative approach for extracting deep features from the MR image named Deep Efficient Reduced Local Derivative Pattern (DERLDP). This approach uses the Efficient Reduced Local Derivative Pattern to extract the deep features using Convolution Neural Network (CNN). The deep features are employed to enhance the brain tumor partition accuracy. So first see the detailed explanation about Efficient Reduced Local Derivative Pattern. In the Efficient Reduced Local Derivative Pattern first take the neighbor values as mentioned in the above figure. And then apply the below formula to find the difference.
PDP= C-N (1)
In the equation 1 C is the center pixel value and N is the neighbor pixel value. And then calculate the direction by using the below formula.
If PDP1 >0 and PDP2 >0 PV = 1
PDP1<0 and PDP2 <0 PV = 2
PDP1 > 0 and PDP2 <0 PV = 3
PDP1 < 0 and PDP 2 >0 PV = 4 (2)
Then calculate the histogram of the pattern description value. Consider these histograms as the feature vector. Compare the query image with the images in the database employed. Based on the best matches retrieve the images from database. And then these features are given into the CNN.
For classification, after extracting the feature, the next step is to partition the brain image based on these retrieved features using supervised segmentation approach. Among various supervised segmentation approaches this work only concentrate the machine learning approaches. From the various machine learning approaches this work takes only deep classifier named Convolution Neural Network.
Under convolution neural network, the CNN based brain tumor segmentation is divided into two phases such as training and testing. In the training phase, preprocessing, feature exaction and classification with Loss function is performed to make a prediction model. Initially, label the training image set. In the preprocessing image resizing is applied to change size of the image. Finally, the convolution neural network is employed for automatic brain tumor classification. In the proposed CNN, we will train only last layer. We don’t want to train all the layers. So, computation time is low meanwhile the performance is high in the proposed automatic brain tumor classification scheme. The loss function is calculated by using gradient descent algorithm. The raw image pixel is mapping with class scores by using a score function. The quality of particular set of parameters is measured by loss function. It is based on how well the induced scores approved with the ground truth labels in the training data. The loss function calculation is very important to improve the accuracy. If the loss function is high, then the accuracy is low. Similarly, the accuracy is high, when the loss function is low. The gradient value is calculated for loss function to compute gradient descent algorithm.
Repeatedly evaluate the gradient value to compute the gradient of loss function. Algorithm for CNN based Classification is shown in Algorithm 2.
Algorithm 2
- Apply convolution filter in first layer
- The sensitivity of filter is reduced by smoothing the convolution filter (i.e.) subsampling
- The signal transfers from one layer to another layer is controlled by activation layer
- Fasten the training period by using rectified linear unit (RELU)
- The neurons in proceeding layer is connected to every neuron in subsequent layer
- During training Loss layer is added at the end to give a feedback to neural network.
RESULT AND DISCUSSION
The assessment of the supervised machine learning segmentation approaches, this paper employs four metrics namely Dice Similarity Coefficient (DSC), Positive Predictive Value (PPV), Jaccard Index (JI) and Sensitivity (SEN) (Bhandar et al. 2020; Wu et al. 2020). The DSC, metric calculates the overlap among the ground truth and the machine learning partition result based on Eq. (3) (Zhao 2016),
DSC = 2TP/(FP+2TP+FN) (3)
Where True Negative (TN) signifies the amount of brain tumor pixels accurately predicted. True Positive (TP) signifies the amount of non-brain tumor pixels accurately predicted. False Positive (FP) signifies to the amount of brain tumor pixels wrongly predicted as non-brain tumor pixels. False Negative (FN) corresponds to the amount of non-brain tumor wrongly predicted as brain tumor pixels. PPV is a calculated of the amount of FP and TP based on Eq. (4),
PPV = TP/(FP+FP) (4)
The Jaccard Index is calculated as the amount of the intersection of the brain tumor and non-brain tumor pixels divided by the amount of their combination based on Eq. (5),
JI = TP/(TP+FP+FN) (5)
At last, Sensitivity metric is calculated to estimate the number of TP and FN based on Eq. (6),
SEN=TP/(TP+FN) (6)
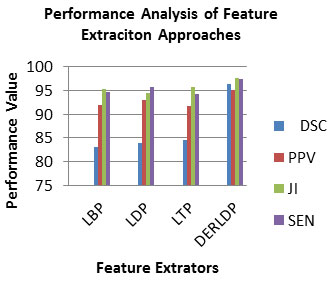
Experiment No 1: Analysis of Feature Extraction Approaches: In this experiment, we will evaluate the contribution of each feature retrieval techniques which are employed in the work. To assess the efficiency of this feature retrieval scheme, the PPV, DSC, JI and SEN measures are employed. It is shown in equation 3,4,5 and 6 correspondingly. Ideally, an excellent feature retrieval approach is accepted to have a high PPV, DSC, JI and SEN value. Table 1 lists the PPV, DSC, JI and SEN measures of feature retrieval approaches.
Table 1. Analysis of PPV, DSC, JI and SEN of BRATS 2015 Dataset for Feature Extraction Approaches
| Feature Descriptor | Dataset | |||
| Metrics |
DSC |
PPV | JI | SEN |
| LBP | 83.093 | 91.993 | 95.213 | 94.633 |
| LDP | 84.003 | 93.013 | 94.383 | 95.723 |
| LTP | 84.523 | 91.603 | 95.603 | 94.203 |
| DERLDP | 96.333 | 94.973 | 97.633 | 97.393 |
As observed from Table 1, the PPV, DSC, JI and SEN of the DERLDP in range 95-97, which is superior than that of the traditional individual feature retrieval method. So, the combined features are best for the brain tumor detection approach. Fig.3 depicted the PPV, DSC, JI and SEN measures of feature retrieval approaches (Bhandar et al. 2020; Wu et al. 2020). As observed from Fig.3, the PPV, DSC, JI and SEN of the DERLDP in range 95-96, which is superior than that of the traditional individual feature retrieval method. So, the combined features are best for the brain tumor detection approach.
Figure 3: Analysis of PPV, DSC, JI and SEN of BRATS 2015 Dataset for Feature Extraction Approaches
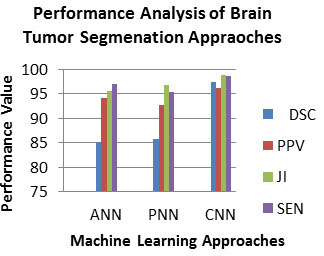
Experiment No 2: Analysis of Brain Tumor Segmentation Approaches: In this experiment, this work will evaluate the contribution of each brain tumor segmentation approaches which are employed in the work. To assess the efficiency of this brain tumor partition technique, the PPV, DSC, JI and SEN measures are employed. It is shown in equation 3,4,5 and 6 correspondingly. Ideally, a good brain tumor segmentation approach is expected to have a high PPV, DSC, JI and SEN value. Table 2 lists the PPV, DSC, JI and SEN measures of brain tumor segmentation approaches (Bhandar et al. 2020; Wu et al. 2020).
Table 2. Analysis of PPV, DSC, JI and SEN of BRATS 2015 for Brain Tumor Segmentation Approaches
| Dataset | ||||
| Machine Learning Segmentation | ||||
| Metrics | DSC | PPV | JI | SEN |
| ANN | 85.212 | 94.222 | 95.592 | 96.932 |
| PNN | 85.732 | 92.812 | 96.812 | 95.412 |
| CNN | 97.542 | 96.182 | 98.842 | 98.602 |
As observed from Table 2, the PPV, DSC, JI and SEN of the CNN in range 96-97, which is superior than that of the other machine learning brain tumor segmentation approach. So, the CNN is best for the brain tumor detection approach. Fig.4 depicted the PPV, DSC, JI and SEN measures of brain tumor segmentation approaches.
Figure 4: Analysis of PPV, DSC, JI and SEN of BRATS 2015 for Brain Tumor Segmentation Approaches
As observed from Fig.4, the PPV, DSC, JI and SEN of the CNN in range 96-98, which is superior than that of the other machine learning brain tumor segmentation approach. So, the CNN is best for the brain tumor detection approach. The contribution of proposed method is evaluated with the existing works. To evaluate the performance of this proposed approach, the performance metrics used are DSC, sensitivity and specificity. Ideally, a good proposed approach is expected to have a high DSC and sensitivity. The proposed method shows 96.33% DSC and 96.32% sensitivity for BRATS data set, which is superior than that of the existing works (Bhandar et al. 2020; Wu et al. 2020).
CONCLUSION
Segmenting the brain tumor is a complicated work, so the error can result in much more. This paper presented a novel deep feature retrieval approach DERLDP and deep classifier method CNN for brain tumor segmentation. The performance of this approach is estimated by PPV, DICE, Sensitivity and Jaccard. The experiments are done on BRATS 2015 dataset. This paper concluded that DERLDP with CNN method produce best result than other existing approaches. In accordance with DICE and Jaccard metrics, this paper justified that the results from DERLDP with CNN segmentation were extraordinarily like to the ground truth segmentation. With the aim of improve CNN, this paper intends to expand the no of training images as well as build up an efficient feature retrieval technique to improve the efficiency of the CNN. In this work, in certain cases, some non-tumor slices were incorrectly classified as tumor (false positive outputs). To improve the accuracy, it is possible to remove the percentage of the image (region of the skull), in the future work and thereby reducing the possibility of false positives in areas that are certainly not. The system will also be experimented in future use of 3D brain datasets.
REFERENCES
Abhishta Bhandari., Jarrad Koppen and Marc Agzarian (2020). Convolutional neural networks for brain tumour segmentation, Insights into Imaging volume 11, Article number: 77.
Ahmmed, R., Swakshar, A.S., Hossain, M.F. and Rafiq, M.A., (2017). Classification of tumors and its stages in brain MRI using support vector machine and artificial neural network. In 2017 International Conference on Electrical, Computer and Communication Engineering (ECCE) (pp. 229-234). IEEE.
El-Dahshan, E.S.A., Mohsen, H.M., Revett, K. and Salem, A.B.M., (2014). Computer-aided diagnosis of human brain tumor through MRI: A survey and a new algorithm. Expert systems with Applications, 41(11), pp.5526-5545.
Hemanth, D.J., Vijila, C.K.S., Selvakumar, A.I. and Anitha, J., (2014). Performance improved iteration-free artificial neural networks for abnormal magnetic resonance brain image classification. Neurocomputing, 130, pp.98-107.
Hinton, G.E., Osindero, S. and Teh, Y.W., (2006). A fast learning algorithm for deep belief nets. Neural computation, 18(7), pp.1527-1554.
Jayadevappa, D., Srinivas Kumar, S. and Murty, D.S., (2011). Medical image segmentation algorithms using deformable models: a review. IETE Technical review, 28(3), pp.248-255.
Jiang, J., Trundle, P. and Ren, J., (2010). Medical image analysis with artificial neural networks. Computerized Medical Imaging and Graphics, 34(8), pp.617-631.
Kamalaveni, V., Rajalakshmi, R.A. and Narayanankutty, K.A., (2015). Image denoising using variations of Perona-Malik model with different edge stopping functions. Procedia Computer Science, 58, pp.673-682.
Krizhevsky, A., Sutskever, I. and Hinton, G.E., (2012). Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems (pp. 1097-1105).
LeCun, Y., Kavukcuoglu, K. and Farabet, C., (2010). Convolutional networks and applications in vision. In Proceedings of 2010 IEEE international symposium on circuits and systems (pp. 253-256). IEEE.
Lei, J., Li, G., Zhang, J., Guo, Q. and Tu, D., (2016). Continuous action segmentation and recognition using hybrid convolutional neural network-hidden Markov model. IET Computer vision, 10(6), pp.537-544.
Logeswari, T. and Karnan, M., (2010). An improved implementation of brain tumor detection using segmentation based on hierarchical self-organizing map. International Journal of Computer Theory and Engineering, 2(4), p.591.
Machhale, K., Nandpuru, H.B., Kapur, V. and Kosta, L., (2015). MRI brain cancer classification using hybrid classifier (SVM-KNN). In 2015 International Conference on Industrial Instrumentation and Control (ICIC) (pp. 60-65). IEEE.
Madabhushi, A. and Lee, G., (2016). Image analysis and machine learning in digital pathology: Challenges and opportunities.
Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, R. and Lanczi, L., (2014). The multimodal brain tumor image segmentation benchmark (BRATS). IEEE transactions on medical imaging, 34(10), pp.1993-2024.
Mohsen, H., El-Dahshan, E.S.A., El-Horbaty, E.S.M. and Salem, A.B.M., (2018). Classification using deep learning neural networks for brain tumors. Future Computing and Informatics Journal, 3(1), pp.68-71.
Pan, Y., Huang, W., Lin, Z., Zhu, W., Zhou, J., Wong, J. and Ding, Z., (2015). Brain tumor grading based on neural networks and convolutional neural networks. In 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (pp. 699-702). IEEE.
Şentaş, A., Tashiev, İ., Küçükayvaz, F., Kul, S., Eken, S., Sayar, A. and Becerikli, Y., (2020). Performance evaluation of support vector machine and convolutional neural network algorithms in real-time vehicle type and color classification. Evolutionary Intelligence, 13(1), pp.83-91.
Singh, A., (2015). Detection of brain tumor in MRI images, using combination of fuzzy c-means and SVM. In 2015 2nd International Conference on Signal Processing and Integrated Networks (SPIN) (pp. 98-102). IEEE.
Singh, G. and Ansari, M.A., (2016). Efficient detection of brain tumor from MRIs using K-means segmentation and normalized histogram. In 2016 1st India International Conference on Information Processing (IICIP) (pp. 1-6). IEEE.
Sudharani, K., Sarma, T.C. and Rasad, K.S., (2015). Intelligent Brain Tumor lesion classification and identification from MRI images using k-NN technique. In 2015 International Conference on Control, Instrumentation, Communication and Computational Technologies (ICCICCT) (pp. 777-780). IEEE.
Suk, H.I., Lee, S.W., Shen, D. (2017). Alzheimer’s Disease Neuroimaging Initiative and deep ensemble learning of sparse regression models for brain disease diagnosis. Medical image analysis, 37, pp.101-113.
Wentao Wu, Daning Li, Jiaoyang Du, Xiangyu Gao, Wen Gu, Fanfan Zhao, Xiaojie Feng, Hong Yan, (2020) An Intelligent Diagnosis Method of Brain MRI Tumor Segmentation Using Deep Convolutional Neural Network and SVM Algorithm”, Computational and Mathematical Methods in Medicine, vol. 2020, Article ID 6789306.
Yan, C., Coenen, F. and Zhang, B., (2016). Driving posture recognition by convolutional neural networks. IET Computer Vision, 10(2), pp.103-114.
Yan, C., Xie, H., Yang, D., Yin, J., Zhang, Y. and Dai, Q., (2017). Supervised hash coding with deep neural network for environment perception of intelligent vehicles. IEEE transactions on intelligent transportation systems, 19(1), pp.284-295.
Yazdani, S., Yusof, R., Karimian, A., Pashna, M. and Hematian, A., (2015). Image segmentation methods and applications in MRI brain images. IETE Technical Review, 32(6), pp.413-427.
Zeineldin, R.A., Karar, M.E., Coburger, J. (2020) DeepSeg: deep neural network framework for automatic brain tumor segmentation using magnetic resonance FLAIR images. Int J CARS 15, 909–920.
Zhao, L. and Jia, K., (2016). Multiscale CNNs for brain tumor segmentation and diagnosis. Computational and mathematical methods in medicine.