1Department of Biomedical Science, SDM Research Institute for Biomedical Sciences, (Shri Dharmasthala Manjunatheshwara University- A State Private University), 5th Floor, Specialty Block, SDMCMS&H, campus, Manjushree Nagar, Sattur-Dharwad-580009, Karnataka-India
2Department of Microbiology/ FST, GITAM Institute of Science, GITAM (Deemed to be University)
Gandhi Nagar, Rushikonda, Visakhapatnam-530045 Andhra Pradesh – India.
3VRDL Laboratory, Department of Microbiology, Hassan Institute of Medical
Science (HIMS), Hassan-573201, Karnataka- India.
4Department of Biotechnology, Gulbarga University Kalaburagi- 584102, Karnataka- India.
Corresponding author email: ckelmani@gmail.com
Article Publishing History
Received: 11/07/2020
Accepted After Revision: 20/09/2020
Vancomycin resistant Enterococcus faecalis is concern for both public health and medical importance associated with serious for multidrug resistant infectious and also resistant for last resort antibiotic. It is an opportunistic pathogen which represents one of the agents of nosocomial infection in hospitalized patient. The aim of the present study was to determine the cell wall thickness by transmission electron microscopy and uncovering of resistant gene responsible gene for the vancomycin resistance E. faecalis strains isolated by clinical samples from Gulbarga region. Isolation and identification of E. faecalis of clinical isolates were done by using standard culturing and screening protocol. Antibiotic susceptibility and MIC were determined by using disc diffusion and broth dilution method. The thickness of cell wall of the stains was study by using TEM and vanA gene was detected by Polymerase Chain reaction.
In our study among 76 isolates of E. faecalis, 12 strains were has showed high resistance to gentamycin and vancomycin antibiotics. TEM – analysis showed cell wall and increased sepate as compared to the normal E. faecalis 122 strain and standard culture E. faecalis NCIM 5625. The VREF 122 strain has showed amplified product size of 352 pb of vanA gene. The study on TEM provides better evaluation in cell wall thickness treated with vancomycin antibiotic and PCR for the detection of resistant gene. These findings indicated the cell wall thickness is a characteristic phenotype of E. faecalis exposed to vancomycin antibiotic and PCR reveals the presence of the vanA gene. It is therefore believed that cell wall thickness and detection of resistance gene plays an important role in mechanism of action of Vancomycin antibiotic. Further these tools may find use in markup application employing detection of resistance pattern in the bacteria.
E. faecalis, TEM, vanA gene, MDR, VRE, cell wall thickness
Oli A. K, Shivshetty N, ManjunathChavadi, Narasanna R, Kelmani R. C. Transmission Electron Microscopy Study of Multidrug Resistant of Enterococcus faecalis and amplification of vanA Gene in Clinical Samples. Biosc.Biotech.Res.Comm. 2020;13(3).
Oli A. K, Shivshetty N, ManjunathChavadi, Narasanna R, Kelmani R. C. Transmission Electron Microscopy Study of Multidrug Resistant of Enterococcus faecalis and amplification of vanA Gene in Clinical Samples. Biosc.Biotech.Res.Comm. 2020;13(3). Available from: https://bit.ly/396Pr7B
Copyright © Oli et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Enterococci are commensal organism that acts as opportunistic pathogens. Currently, they have been studied to be among leading pathogens of nosocomial infections, and thus they are a major international health burden (Dulber et al., 2020).Among the enterococci, Enterococcus faecalis cause 80-90 % of infections and it is a gram-positive commensal member of the gut microbiota of wide range organisms. With the advent of antibiotic therapy, it has emerged as multidrug resistant, hospital acquired pathogen (Daria et al., 2013). It majorly causes in humans such as bacteremia, endocarditis, intra-abdominal, pelvic infections and urinary tract infections. However, inappropriate vancomycin use has resulted in the emergence of Vancomycin resistant enterococci (VRE) and capably of transferring the resistance gene to another organism. VRE infection can be acquired through colonization with VRE or hospital environmental (Jatapat et al., 2019). Enterococci gladly accrue mutations and exogenous genes that confer additional resistance. They develop resistance to vancomycin by exchange of genetic material among themselves and or with another genera. The enterococci may acquire resistance through van associated genetic elements like vanA, vanB, vanD, vanE, vanG, Van L of which vanA and vanB are the most prevalent genotypes in clinical isolates (Addisu et al., 2020).
In Asian countries the prevalence was decreased may due to recent emergence of this resistance in this continent and only a handful of studies to document. The prevalence of VRE in India has been reported the highest was in New Delhi with 23%, Chandigarh 8% and lowest in Mumbai with 5.5% respectively and all of vanB phenotype study reported by Manimala et al., (2019).Transmission electron microscopy will insightful impact on understanding of bacteria and other microbial populations, it provides the 1000 fold improvement in resolution and allows to find the minor changes in topological and structure of bio-membrane organelles. (Shruthi and Vivek 2017).It is important to study the prevalence, pattern of resistance especially in this region of South India which has very few reports on Enterococcus faecalis. The present objective of the study to better understand the antimicrobial action and changes in cell wall thickness induced by vancomycin in E. faecalis.
MATERIAL AND METHODS
Isolation of Enterococcus faecalis: The samples were examined for the presence of pathogenic. In the present study bile esculin azide agar was used for primary isolation of E. faecalis. Microscopic observation was done for confirmation and subculture again 3 to 4 times to isolate pure culture of E. faecalis. The single isolated colony was inoculated in BHI broth and incubated at 37°C for 18 hrs, glycerol (30%) were used for preparation of stock culture and stored in -80 °C for further use.
Antimicrobial Susceptibility testing: The standard disk diffusion method was used to perform antimicrobial susceptibility testing on Muller Hinton agar (Hi-Media, India) as recommended by the Clinical and Laboratory Standards Institute (CLSI 2007). The most commonly used antibiotics used for the tests were vancomycin, ampicillin, oxacillin, rifamycin, ciprofloxacin, tobramycin, gentamycin, teicoplanin and streptomycin.
Determination of minimal inhibitory concentration (MIC’s) of Vancomycin and Gentamycin by broth dilution method:MIC for antibiotic (i.e, Gentamycin and Vancomycin) concentration ranging from 6-256 μg/ml and 512 to 1026μg/ml respectively were prepared with BHI broth and used to test each isolate and final concentration of each antibiotic were 10 mg/ml.
Transmission Electron Microscopy:TEM was performed for the morphological characterization of the cell wall ultra-structure of GREF and VREF isolates. The protocol for the preparation and examination was followed as described earlier (Hanaki et al., 1998). The overnight culture was centrifuge at 8000 rpm for 10 mins 4º C, the supernatant fluids were discarded, and the cell pellets were washed twice with 50 mM potassium phosphate buffer (pH 7.0). Bacteria were then fixed in 3 % glutaraldehyde (in 0.1 M sodium phosphate buffer, pH 7.2). Then followed bacteria were fixed with 3% glutaraldehyde for overnight at 4º C. For high contrast amplification, the bacterial cells were treated with 2 % uranyl acetate in 95 % alcohol for 1 h at 20º C in the dark. Infiltration is done with help of araldite propylene oxide (1:1) solution for five times. Cells were subjected to dehydration with 70 % ethanol for 18 hrs in acrylic resin. The ultra-microtome was used to prepare ultra-thin section, finally stained with lead citrate and uranyl acetate and examined under Transmission electron (Techn G2 Spirit TEM, at 80 KV).
Plasmid DNA Analysis:The plasmid isolation kit (Bangalore gene pvt.ltd) were used for the extraction of Plasmid DNA from vancomycin resistant isolates by user manual of kit, further it was analyzed in 0.8% agarose gel electrophoresis containing 5 μg/mL of ethidium bromide at 3.5 V/ cm for 4 h in a Tris EDTA buffer (TAE). Molecular markers were used as a size reference for molecular determinations.
vanA Gene Amplification:Plasmid DNA from vancomycin resistant enterococci isolated by the plasmid isolation kit (Bangalore gene pvt. ltd) were subjected to amplification assays employing the oligonucleotide forward primer (5’-GGGAAAACGACAATTGC-3’) and reverse primer (5’-GTACAATGCGGC CGTTA-3’) and amplification conditions as described by (Dutka et al.,1995). The PCR reaction consisted of Initial denaturation at 94 ˚C for 10 mins, followed by 35 cycles of denaturation to 94˚C for 30 secs, annealing to 47 ˚C for 45 secs, then extension to 72˚C for 30 secs. Followed by final extension at 72 ˚C for 10 mins. The amplified products were resolved by electrophoresis on a 1% Agarose–TAE gel containing gel red.
RESULTS AND DISCUSSION
Bacterial Isolates and Antimicrobial Susceptibility Test: In the present study, strains of E. faecalis were isolated from clinical samples A total of 250 samples were collected, among them 122 have showed positive for different clinical isolates. The E. faecalis 76 (62.29 %) was the predominant isolates from urine, pus, CSF and blood samples. The E. faecalis isolates were Gram positive. The biochemical characteristics for the all isolates were positive for tellurite reduction and arginine hydrolysis, arabinose, raffinose and mannitol expect for catalase sorbitol and lactose. The results of the susceptibility tests are carried out by disc diffusion method for 76 E. faecalis strains showed resistant to the different antibiotic like vancomycin (77.63%), gentamycin (64.47%) and oxacillin (55.26%) antibiotics, and were multi drug resistant. The isolates were found sensitive to rifamycin (61.84%), teicoplanin (55.26%) streptomycin 52.63%) and tobramycin (51.13%).
Determination of MIC’s in E. faecalis isolates:MIC’s for gentamycin among 76 isolated 12 E. faecalis high resistance to gentamycin and Vancomycin among them 5 isolates showed ≥1024 μg/ml and 5 isolates had MIC of ≥512 μg/ml and 2 strains showed 256 μg/ml. The vancomycin MIC for 8 isolates showed ≥64 μg/ml and 4 isolates had ≥ 128 μg/ml of the total 12 isolates were tested as shown in the Table.1.
Table 1. MICs determination of Gentamycin and Vancomycin resistance of E. faecalis Isolates
| Isolates | Sample | MIC μg/ml | |
| Gentamycin | Vancomycin | ||
| 121 | Urine | 1024 | 128 |
| 122 | Urine | 1024 | 128 |
| 123 | Urine | 1024 | 128 |
| 124 | Urine | 1024 | 64 |
| 125 | Urine | 1024 | 64 |
| 136 | Blood | 512 | 64 |
| 137 | Blood | 512 | 64 |
| 138 | Blood | 512 | 64 |
| 139 | Blood | 512 | 64 |
| 140 | Blood | 512 | 64 |
| 141 | CSF | 256 | 128 |
| 142 | Pus | 256 | 64 |
Transmission Electron microscopy:TEM was employed to examine the cell morphology (especially cell wall thickness) of E. faecalis in in-vitro using gentamycin and vancomycin. Using broth dilution method, E. faecalis (GREF 122 and VREF 122) was adapted during MIC study and further sub cultured and stored at -80º C (expressed high MICs, ≥1024 µg/ml and 128 µg/ml). To ensure the resistance mechanism of E. faecalis to gentamycin and vancomycin, the TEM was carried out along with control isolates (EF122 and NCIM 5025). The isolates showing baseline resistance of 16 µg/ml MIC were subjected to chemical fixation prior to examination.
Table 2. Averaged means values of Cell wall thickness of the isolates
| Name of the isolates | Measurement of thickness magnification at X 68,000 nm | |
| Cell wall | Septum | |
| Control Strain EF 122 | 30.598 nm | 18.902 nm |
| Standard Culture
E. faecalis NCIM 5025 |
30.832 nm | 24.358 nm |
| Antibiotic treated (Gentamycin) Strain EF 122 | 40.164 nm | 35.770 nm |
| Antibiotic treated (Vancomycin) Strain EF 122 | 47.842 nm | 46.358 nm |
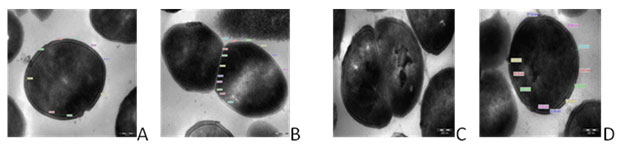
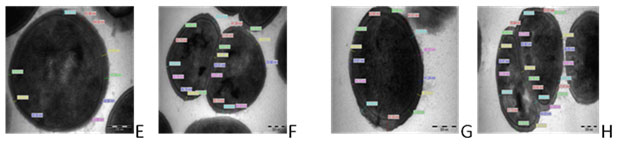
TEM photographs showed that there was significant alteration in the thickness of the bacterial cell wall. The control strain EF 122 showed the cell wall thickness of 30.598 nm and septum of 18.902 nm with X68,000 magnification as shown in Fig.1 a & b. The standard strain NCIM E. faecalis 5025 showed 30.832 nm cell wall thickness and septum of 24.358 nm with X68, 000 magnifications as shown in Fig.1 c & d. The isolates when exposed to gentamycin the cell wall and septum thickness was increased in size of 40.164 nm and 35.770 nm respectively at magnification at X68, 000 as show in Fig. 2 e & f. Similarly, the cell wall and septum thickness was increased higher in size of 47.842 nm and 46.358 nm respectively for vancomycin antibiotic as shown in Fig. 2 g & h.
Figure 1: Transmission electron micrographs of E. faecalis (A) Control Isolate E. faecalis EF122 (B) Control of E. faecalis EF122 Septum; (C) NCIM 5205 E. faecalis Std Strain; (D) NCIM 5205 E. faecalis Std. Strain septum magnification at X68, 000.
Figure 2. Transmission electron micrographs of E. faecalis isolates (E) Gentamycin treated faecalis EF122 isolate; (F) Gentamycin treated E. faecalis EF 122 isolate of Septum;(G) Vancomycin treated E. faecalis EF 122 isolate;(H) Vancomycin Treated E. faecalis EF122 isolate septum magnification at X68,000
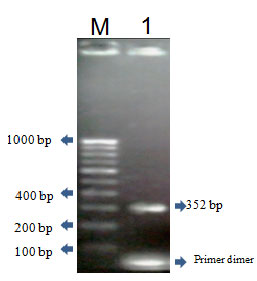
Plasmid DNA analysis and vanA gene amplification: Figure 3. Reports the plasmid profile of human clinical isolates. The two isolates 121 and 122 are independently of their origin had more than one plasmid with different molecular weight. Among the observed plasmid, those ranging from 2 strains, more than one plasmid species were detected in isolates. Among the observed plasmids, those ranging approximately from 21 and 19 kbp in molecular size. (Not data shown) shows an example of amplification of the 352 pb fragment in plasmid DNA of isolates, confirming the affiliation to the vanA gene cluster. This result also demonstrates the extra chromosomal location of the glycopeptide resistance determinants. The amplified PCR product was sequenced and submitted to NCBI (Acc .No JN791694).
Figure 3: PCR amplification of vanA gene Lane M- 100bp ladder and Lane-1 amplified Product vanA gene of E. faecalis EF122
Drug resistant infectious provides opportunities to address the associated clinical and public health burden on individuals, health systems, and society, (Olaniyi et al., 2020). Worldwide there is a trend in increasing of vancomycin resistant enterococci ever since first described in 1987, although the epidemiology of these microorganism varies widely in different geographical areas (Hossein and Mohammad 2014). E. faecalis has recently evolved from a generally a virulent commensal into an multi-drug resistant healthcare-associated pathogen causing difficult-to-treat infections. Therefore, studies of E. faecalis resistance have increased. The most species distribution of the enterococcus species was E. faecalis with (81.72%) isolated, followed by E. faecium (12.9%) as reported by Jahnabi et al., 2016. In recent study they have been reported has (64%) of E. faecalis and E. faecium (36%) in Chennai, India (Alexander et al., 2020).
In our studies E. faecalis occurrence was (62.29%) the difference may be due to geographical variation. Vancomycin resistance was high with numbers of screened isolates were less 28/31 (90.62%) reported in North Indian tertiary care hospital (Hibs et al., 2020). In the year 2016 a study showed high rate (35.3%) of VRE carriage as compared to studies that have estimated comparatively low VRE colonization on admission to the ICU from Europe (2.7%), US (12.3%), South America (7%) and other Asian countries (5.3%) (Prakhar et al., 2016). In our study, about 77.64 % of E. faecalis strains showed high resistant to vancomycin by disk diffusion test and MIC of 12 strains as showed ≥ 64 to ≥128 μg/ml for vancomycin respectively. The last therapeutic resort for enterococci was vancomycin.
Transmission electron microscopy has been invaluable tool in imaging bacteria and their components. The first report to show cell wall thickening was in S. aureus following treatment with acriflavine. Beta-lactam antibiotics which inhibit cell wall synthesis cause marked thickening of the cross wall in Staphyloccocci. MRSA has showed that vancomycin is affinity trapped inside the peptidoglycan layers by false targets, resulting in peripheral wall thickening (Mako and Jun 2009). In present study the isolate vancomycin resistant E. faecalis EF122 showed lesser thickness of cell wall and septum compared standard strain NCIM E. feacalis 5025 when grown in the medium without vancomycin. In contrast, when the isolate exposed to 16 µg/ml of vancomycin and gentamycin increased thickness of cell wall was 9.564 nm and 16.566 nm against gentamycin and vancomycin, whereas in case of septum 26.868 nm and 27.456 nm with gentamycin and vancomycin respectively.
In our study PCR proved helpful in detecting the van genotypes present in this geographic region. The study reveals an expression of vanA gene in the E. faecalis strain with 352bp fragment in the plasmid DNA and it confirms isolates the affiliation to the vanA gene cluster and the extra chromosomal location of the glycopeptide-resistance determinants. The vanA type resistance is dominant in the United States and Europe whereas vanB type resistance is more frequent in Australia and Southeast Asia, (Nerfis and Baris 2020). However, VRE strains carrying vanB or both vanA/vanB have reported in various countries (Coombs et al., 2014, Marcade et al., 2014, Papagiannitsis et al., 2017).In India the most of strains of VRE has showed the vanA genotype (Hiba et al., 2020). In our study, the increase in the rate of prevalence of E. faecalis and emergence of multidrug resistance among them (Ajay et al., 2012), highlights the significance of rapid and accurate in identification of enterococci to the species level for imitating appropriate therapeutic regimen, and reemphasizes the importance of the implementation of appropriate infection control measures to limit the nosocomial spread of these unusual species in any nosocomial.
ACKNOWLEDGMENTS: The first author is grateful to Council Scientific and Industrial research for Senior Research fellowship (CSIR-SRF New-Delhi, India) and enabling him to complete the research work.
Authors Contributions: All authors have equal contribution in bringing out this research work
Conflict of Interest: None
REFERENCES
Addisu M., Chalachew G., Tesfaye A. (2020). Prevalence of Vancomycin resistant enterococci (VRE) in Ethiopia: a systematic review and meta-analysis. BMC Infectious Disease.20:124; 1-12.
Ajay K.O., Rajeshwari H., Nagaveni S., Kelmani C.R. (2012). Antimicrobial susceptibility pattern of enterococcus species isolated from clinical samples in south India. Journal of recent advances in applied sciences. 27; 06-10.
Alexander K., Kesavaram P., Praveen S., Sumathi G., Jeevan M. (2020). Higher incidence of Pristinamycin resistance among Enterococcus faecium with iMLSB / cMLSB Phenotype. Biomedical & Pharmacology Journal. 13(1); 27-31.
Clinical and Laboratory Standard Institute. Performance Standards for antimicrobial susceptibility testing (2007) seventeenth informational supplement.M100-s17. Vol.27 (1).
Coombs G.W., Pearson J.C., Daley D.A. et al. (2014). Molecular epidemiology of enterococcal bacteremia in Australia. J Clin Microbiol. 52; 897–905
Daria V.T., Melissa J.M., Michael S.G. (2013). Structure, Function and Biology of the Enterococcus faecalis cytolysin.Toxins.5; 895-911.
Dubler S., Lenz M., Zimmermann S., Richter D.C., Wesis K.H., Mehrabi A., Mieth M., Bruckner T., Weigand M.A., Brenner T., Heininger A. (2020). Does Vancomycin resistance increase mortality in Enterococcus faecium bacteraemia after orthotopic liver transplantation? A retrospective study. Antimicrobial Resistance and Infection Control. 9:22; 1-10.
Dutka M.S., Evers S., Courvalin P. (1995).Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol.33; 24-27.
Hanaki H., Kuwahara A.K., Boyle V. S, Daum RS, Labischinski H, and Hiramatsu K. (1998). Activated cell wall synthesis is associated with vancomycin resistance in methicillin resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother. 42; 199-209.
Hiba S., Anuradha S., Shariq A., Mohd S. (2020). Emergence of linezolid resistance in enterococci: prevalent genotypes and resistance pattern in vancomycin resistant enterococci in a North-Indian tertiary care hospital. N Z J Med Lab Sci.74; 27-30.
Hossein S.K., Mohammad A. (2014). Vancomycin resistant Enterococcus faecium and Enterococcus faecalis isolated from education hospital of Iran. Maedica: A Journal of Clinical Medicine. 9(4); 323-327.
Jahnabi B., Reema N., Lahari S. (2016). Drug resistance in Enterococcus species in a tertiary level hospital in Assam, India. Indian Journal of Medical Research. 143(1); 107-110.
Jatapat H., Dhitiwat C., Sudaluck T., Wichai S. (2019). Vancomycin-resistant enterococcal infection in a Thai university hospital: clinical characteristics, treatment outcomes, and synergistic effect. Infection and Drug Resistance.12; 2049-2057.
Manimala E., Rejitha I.M., Revathy C. (2019). Detection of Vancomycin resistant enterococci in various clinical sample isolates from a tertiary care Centre. Int J Curr Microbiol App Sci 8(2); 915-921.
Marcade G., Micol J., Jacquier H, et al. (2014). Outbreak in a haematology unit involving an unusual strain of glycopeptide-resistant Enterococcus faecium carrying both vanA and vanB genes. J Antimicrob Chemother. 69(2); 500–505.
Nergis A., Baris O. (2020). Antibiotic resistance and Molecular Epidemiology of vancomcyin resistant enterococci in a tertiary care hospital in turkey. Infection and Drug Resistance.13; 191-198.
Olaniyi A., Niklas W., Annicka R., Tim E., Robby M. (2020). The ongoing challenge of Vancomycin resistant Enterococcus faecium and Enterococcus faecalis in Europe: an epidemiological analysis of blood stream infections. Emerging Microbes and Infections. (9)1; 1180-1193.
Papagiannitsis C.C., Malli E., Florou Z, et al. (2017). First description in Europe of emergence of Enterococcus faecium ST117, carrying both vanA/vanB genes, isolated in Greece. J Glob Antimicrob Resist. 11:68–70.
Prakhar A., Lipika S., Varsha G., Vishal G., Jagdish C.(2016).Vancomycin-resistant enterococci and healthcare-associated risk factors in pediatric intensive care unit. Indian J Med Res.149; 71-73.
Shruti S., Vivek K.B. (2017). Visual demonstration of transmission electron microscopy for intracellular observation of a single bacterial cell. Bangladesh J Pharmacol.12:23-27.