Seri-biotech Research Laboratory, Central Silk Board, Kodathi, Carmelram Post, Bengaluru- 560 035, India.
Corresponding author email: ravikumarpillai@gmail.com
Article Publishing History
Received: 10/04/2020
Accepted After Revision: 10/06/2020
Silk sericin has recently reported to be a promising biopolymer with wide applications in biomedicine and biotechnology. Combining sericin with other functional proteins could enable the development of novel biomaterials. Most of the functional properties of sericin, including its hydrophilicity and mechanical strength are attributed by the internal repetitive 38- amino acid motif of Ser I gene. Because of the unique repetitive sequence of sericin, recombinant expression of it in heterologous expression system is difficult and hence accomplished only in bacteria so far. In this study, we report an attempt to express sericin and sericin-cecropin B fusion using the cell-free and Pichia pastoris expression systems. Vector constructed using both the systems were successful as sericin and sericin cecropin B recombinant proteins were expressed. SDS-PAGE followed by western blot demonstrated that the recombinant sericin-cecropin B protein of approximately 16 and 20 kDa were expressed in cell-free system and Pichia, respectively. However, the yield of the recombinant proteins was comparatively lower than that in bacterial expression system.
Silk, Sericin, Fusion, Expression, Pichia pastoris, Cell-free system
Thomas D. S, Manoharan C, Rasalkar S, Mishra R. K, Gopalapillai R. Expression of Silk Sericin-Cecropin B Fusion Protein in Pichia pastoris and Cell-Free System. Biosc.Biotech.Res.Comm. 2020;13(2).
Thomas D. S, Manoharan C, Rasalkar S, Mishra R. K, Gopalapillai R. Expression of Silk Sericin-Cecropin B Fusion Protein in Pichia pastoris and Cell-Free System. Biosc.Biotech.Res.Comm. 2020;13(2). Available from: https://bit.ly/3amx5hR
Copyright © Thomas et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Silk, the natural fibre with its lustrous appearance has been ruling the textile industry since centuries. The silk proteins, fibroin and sericin, with highly repetitive sequences and secondary β sheet structure exhibits environmental stability, mechanical strength, biodegradability and biocompatibility making them suitable candidates as biomaterial. Both the proteins have proven to be suitable in a range of applications, cosmetics, as dietary supplement, tissue engineering and biomedicine. However, obtaining these proteins from their natural source is a major concern due to drawbacks like batch-to-batch variation and impurities which often limits their applications. In this regard, recombinant silk proteins are desired which can be a source of well characterized, pure proteins that can meet the stringent needs in biomedical applications and also is non-destructive. Further, recombinant expression enables hybridization of the protein with active domains to generate multifunctional proteins, (Holland et al., 2019, Thomas et al., 2020).
Sericin, the unutilised by-product of the textile industry is a mixture of four glycoproteins, generated by differential splicing and secreted in the middle silk glands of Bombyx mori. Among them, Ser 1 encoded by Ser1, consists of repeats of a 38-amino acid motif that contributes towards the hydrophilicity, mechanical strength and other functional properties. In the recent years, sericin has shown potential application as, a supplement in culture media, cryoprotectant, vehicle for drug delivery, metabolic effectors in organic systems, in wound healing and tissue engineering, (Teramotoet al., 2006; Huang et al., 2003,Kundu et al., 2008, Dong et al., 2019).
Recombinantly expressed spider silk (Spidroin) and B. mori silk proteins in general have shown to be functional. Moreover, the chimeric proteins of spidroin and fibroin with enhanced features have also been generated and found applicable in tissue engineering (Aigner et al., 2018). However, reports on successful recombinant expression of sericin have been limited to two reports (Tsujimoto et al., 2001; Huang et al., 2003), using bacterial expression system, but no report of sericin fusion protein. Recently we have recombinantly expressed functional sericin and sericin cecropin fusion proteins in bacteria (Thomas et al., 2020, a & b).
In this study we explore the possibility of expressing these proteins in cell-free and Pichia pastoris expression systems for the first time.
MATERIALS AND METHODS
Vector constructions: The cloning of sericin and sericin cecropin B is described in our earlier papers (Thomas et al 2020 a). Briefly, the PCR product sericin or sericin-cecropin B fusion flanking SgfI and PmeIwere introduced in to pF25A ICE T7 Flexi vector(Promega) having the corresponding restriction sites, generating plasmids pS25A and pSC25A, respectively. For P. pastor is expression system, sericin alone and sericin-cecropin B fusion were amplified using the above plasmid as template with compatible primers flanking restriction sites (MlyI, KpnI and FseI) for insertion to pPink-LC vector (Invitrogen). The primers were as follows: SerFw- 5’ GCGAGTCGCACTAGCACAGACCTGGCAGGATCCAGTACA 3’, SerRev- 5’ GGGGTACCTTAATGGTGATGATGGTGATGACTATATCCTTCAGTGCTGCTACCAGC3’ and CecRev- 5’ CGGCCGGCCTTAATGGTGATGATGGTGATGTTTTCCTATAGCTTTAGCCGAACC 3’. Sericin sense primer was designed to create fusion product with Saccharomyces cerevisiae α-mating factor pre-sequence (with flanking 5’ EcoRI and 3’ MlyI compatible sites) for secreted expression. In both the constructs, a flanking region encoding six histidines followed by a stop codon was added at 3’end. All constructs were verified by DNA sequencing.
Cell-free expression:The reactions were set up in 1.5 ml as per manufacturer’s protocol. The plasmids, pS25A and pSC25A, served as templates for in vitro transcription and translation. Individual reactions were setup using, 40 μl TNT T7 ICE master mix, 4 μg plasmid DNA template and milli Q to a final volume of 50 μl. Reactions were gently mixed and incubated at 28oC for 4 h and analysed.
Pichia pastoris expression: The plasmids were linearized and used to transform P. pastoris (PichiaPink, strain 2, Invotrogen) using the Pichia Easy Comp protocol according to the manufacturer’s instructions. After transformation the entire mixture was spread onto Pichia adenine dropout (PAD) selection plates and incubated at 30oC for 3 to 10 days, until distinct colonies were observed. The expression vector without any insert was also transformed and served as control. The transformants (white colonies) were picked and the plasmid integration in the yeast genome was confirmed by PCR. Single colonies were inoculated into 2.5 ml of BMGY medium (Buffered Glycerol-complex medium, pH 6.0, Invitrogen) and incubated at 30oC with vigorous shaking until it attained log phase growth (A600= ~3). These cultures were used to inoculate 50 ml of BMGY and cells were further incubated at 30oC with vigorous shaking until A600 reached 3.0. To induce expression, cultures were centrifuged at 1,500 x g for 5 min at room temperature and the cell pellets were resuspended in 10 ml BMMY medium (Buffered Methanol-complex medium, pH 6.0, Invitrogen). The cultures were incubated at 22oC with vigorous shaking continuously till 96 h, with additions of methanol (1%) every 24 h. Cells were harvested every 24 h by centrifugation at 1,500 x g for 5 min. The supernatant and pellets were separated and stored at -80oC till further processing. The cell pellets and supernatants containing recombinant proteins were processed according to the manufacturer’s instruction.
Western blot: Purified protein samples were separated on a 15% SDS-PAGE and electrophoretically transferred (TE77X, Hoefer) onto polyvinylidene difluoride membranes (Hybond, GE). The His6-tagged proteins were probed using mouse anti-histidine primary antibody (1:5000). The proteins were visualizedusing alkaline phosphatase conjugated goat anti-rabbit IgGand 4-nitro blue tetrazolium/5-bromo-4- chloro-3-indolyl phosphate (Western MAX, Amresco).
RESULTS AND DISCUSSION
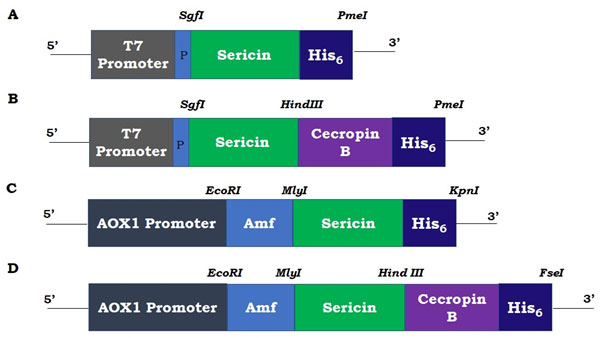
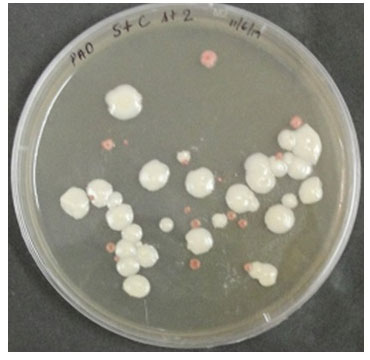
The approximately 300 bp sericin amplicon (including flanking sites) of the Ser 1 geneand the111 bp cecropin B gene were cloned into the pF25A and pPink-LC vectors under T7 and AOX1 promoters, respectively, for cell-free and Pichia expression systems (Figure 1 A-D). All the inserts were confirmed by colony PCR and sequencing and found in-frame with c-terminal histidine tags. Transformation of the Pichia strain 2 with linearized plasmids resulted in white colonies with a few pink ones (Figure2). The chromosomal integration of the heterologous expression cassettes in P. pastoris was confirmed by PCR using genomic DNA as template (Data not shown). These results show the error-free vector constructions and successful integration of the insert into Pichia genome.
Figure 1: Expression vector designs for sericin and sericin-cecropin B. A & B: Cell-free expression plasmid, pF25A, C & D: Pichia pink, pPink-LC, P: polyhedron 5’UTR, Amf: yeast alpha mating factor.
Figure 2: Colonies of Pichia pastoris Pink strain 2 transformed with linearized pPink-LC having sericin-cecropin B on PAD agar plate. White colonies represent cells transformed with recombinant plasmid and dark pink represent colonies without inserts.
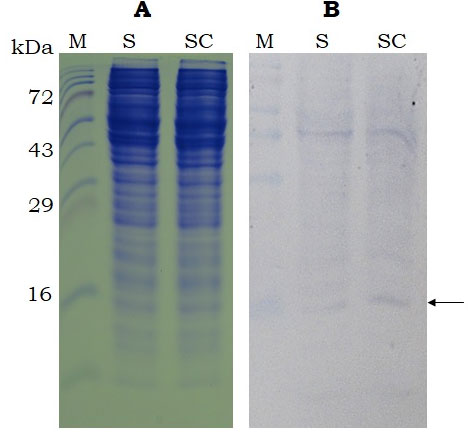
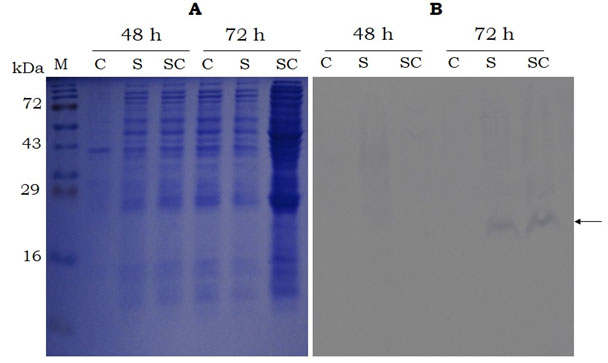
SDS-PAGE and western analysis show the expression of sericin and sericin-cecropin B fusion proteins in both the expression systems. As observed in figure 3B, the western blot analysis of recombinant proteins expressed using cell-free expression system showed band at ~ 16 kDa for both sericin and sericin-cecropin B. However, the expression of recombinant proteins was not visible in the SDS-PAGE analysis. The effect of secreted expression (proteins extracted from culture supernatants) was analysed for various time points up to 72 h in Pichia. As indicated in figure 4B, the candidate protein bands were not visible up to 48 h but visible only at 72 h post-induction. No bands were visible after 72 h (Data not shown). Protein extracted from cells did not show the presence of recombinant proteins at any time points (Data not shown). Western blot showed a band of approximately 20 kDa for both proteins at 72 h. No band was observed in control having proteins extracted from Pichia transformed with empty vector.
Figure 3: SDS-PAGE (15%, A) and Western-blot (B, using anti-histidine antibody) analysis of the recombinant sericin and sericin-cecropin B proteins expressed using cell-free expression system. Proteins were collected after 4 h of incubation. M- Protein marker (Puregene), S- sericin, SC- sericin-cecropin B. Arrow indicates target band of ~ 16kDa.
Figure 4: SDS-PAGE (15%. A) and Western-blot (B, using anti-histidine antibody) analysis of the sericin and sericin-cecropin B proteins secreted by P. pastoris(strain 2) in to the growth medium after 48 and 72 h of induction. M- Protein marker (Puregene), C- control (proteins from P. pastors harbouring empty vector), S- sericin, SC- sericin-cecropin B. Arrow indicates target band of ~ 20kDa.
The theoretical molecular masses and pI for sericin and sericin-cecropin protein are 9.77, 9.87 (4.73, 4.93) and 14.23, 14.04 (6.57, 6.96), respectively, in cell-free and Pichia vector constructs. The apparent molecular mass as noted with western blots is slightly higher than that of the calculated molecular masses in both sericin and sericin-cecropin proteins. This may be due to the unique repetitive amino acid sequence of sericin and pI of sericin and cecropin resulting in reduced electrophoretic mobility. This is in agreement with reports on recombinant sericin expressed in bacteria by Tsujimotoet al., 2001; Huanget al., 2003 and our study (Thomaset al., 2020a). Similarly, a reduction in electrophoretic mobility was observed with genetically engineered spider silk-dentin matrix protein 1 expressed in bacteria (Huang et al., 2007). The reason for the high molecular mass of recombinant proteins in Pichia may be due to the post-translational modifications, especially glycosylation. Compared to the bacterial expression of sericin and sericin-cecropin B (~1.25 mg/L, Thomas et al., 2020).
The level of expression of both proteins was much lesser in the present study as observed by faint bands in western blots. Unlike cell-free system, which has the limitation of producing recombinant proteins in large quantity, Pichia expression system can be improved by optimising culture conditions and use of fermenter. Hence, further work is required for high yield of recombinant sericin or its fusion proteins in P. pastoris expression system. The production of recombinant spider silk proteins using various heterologous systems, including P. pastoris have been reported. The synthetic gene DP-1B, based on the partial amino acid sequence patterns of the major dragline silk protein spidroin 1 and functionalized spider silk fusion protein Z-4RepCT, were expressed in P. pastoris (Fahnestock and Bedzyk, 1997; Jansson et al., 2016).
Further, the in vitro translation of synthetic genes based on MaSp 2, spider dragline silk protein, using E. coli S30 extract have also been reported (Lewis et al., 1996). However, till date there is no report on recombinant expression of sericin or its fusion proteins in P. pastoris or cell-free systems. Our results show the possibility of recombinant expression of difficult-to-express proteins like sericin or its fusion in heterologous expression systems.
ACKNOWLEDGEMENTS
DST, CM, and SR are thankful to Central Silk Board (CSB), Bengaluru, India for financial assistance in the form of Junior and Senior Research Fellowships. This work was supported by a grant from CSB to RG.
REFERENCES
Aigner TB, DeSimone E and Scheibel T (2018) Biomedical applications of recombinant silk-based materials. Adv Mater, 30:1–28
Dong Z, Guo K, Zhang X, Zhang T, Zhang Y, Ma S, Chang H, Tang M, An L, Xia Q and Zhao P (2019) Identification of Bombyx mori sericin 4 protein as a new biological adhesive. Int J BiolMacromol, 132:1121-1130
Fahnestock SR and Bedzyk LA (1997) Production of synthetic spider dragline silk protein in Pichia pastoris. ApplMicrobiolBiot, 47:33-39
Holland C, Numata K, Kovacina JR and Seib FP (2019) The biomedical use of silk: Past, present and future. Adv Healthc Mater, 8:1-26
Huang J, Valluzzi R, Bini E, Vernaglia B and Kaplan DL (2003) Cloning, expression and assembly of sericin-like protein. J Biol Chem, 278:46117-46123
Huang J, Wong C, George A and Kaplan DL (2007) The effect of genetically engineered spider silk-dentin matrix protein 1 chimeric protein on hydroxyapatite nucleation.Biomaterials, 28:2358–2367
Jansson R, Lau CH, Ishida T, Ramstrom M, Sandgren M and Hedhammar M (2016) Functionalized silk assembled from a recombinant spider silk fusion protein (Z-4RepCT) produced in the methylotrophic yeast Pichia pastoris. Biotechnology Journal, 11:687–699
Kundu SC, Dash BC, Dash R, Kaplan DL (2008) Natural protective glue protein , sericin bioengineered by silkworms: Potential for biomedical and biotechnological applications. Prog Polym Sci, 33:998–1012
Lewis RV, Hinman M, Kothakota S and Fournier J (1996) Expression and purification of a spider silk protein: A new strategy for producing repetitive proteins, Protein ExpresPurif, 7:400–406
Teramoto H, Kakazu A and Asakura T (2006) Native structure and degradation pattern of silk sericin studied by 13C NMR spectroscopy Macromolecules, 39:6–8
Thomas DS, Manoharan C, Rasalkar S, Mishra RK and Gopalapillai R (2020) Recombinant sericin-cecropin B fusion protein aids in the proliferation and cryopreservation of human dermal fibroblast cells. Bios Biotech Res Comm 2020 (in press)
Tsujimoto K, Takagi H, Takahashi M, Yamada H and Nakamori S (2001) Cryoprotective effect of the serine-rich repetitive sequence in silk protein sericin. J Biochem, 129:979-986