Ho Chi Minh City University of Food Industry 140 Le Trong Tan Street,
Tan Phu district, Ho Chi Minh City 700000, Vietnam.
Corresponding author email: thehv@hufi.edu.vn
Article Publishing History
Received: 10/03/2021
Accepted After Revision: 28/05/2021
Several species in Panax genus have been identified and utilized as important traditional medicines in Vietnam for decades. Due to the varying degrees of scarcity and value of medicinal herbs, different Panax types are often blended to provide illegal benefits. Current morphology-based identification of Panax species is low accuracy. In this study, Internal Transcribed Spacer (ITS) regions of 23 Panax samples in Vietnam were amplified and sequenced. The obtained sequences were then searched for homology in NCBI Genbank to identify Latin names. The genetic relatedness of ITS sequences were then analyzed by phylogenetic analysis. The results show that 23 studied samples belong to four Panax species. The sequence alignment also reveals the distinct variation in ITS regions of Panax species in Vietnam in the comparison to ITS sequences of Panax species from other countries. The obtained results show high effectiveness of using ITS in distinguishing Panax species in Vietnam. Furthermore, the variation within this region could be developed into specific marker for authenticating ginseng-derived products.
DNA Barcode, Ginseng, Identification, ITS, Panax Species.
Nguyen M. P, Ngo T. K. A, Le T. T, Huynh N. T, Huynh T. T, Le N. T. A, Ho V. T. Utilization of ITS-Based DNA Barcode for Classification of Different Panax Species in Vietnam. Biosc.Biotech.Res.Comm. 2021;14(2).
Nguyen M. P, Ngo T. K. A, Le T. T, Huynh N. T, Huynh T. T, Le N. T. A, Ho V. T. Utilization of ITS-Based DNA Barcode for Classification of Different Panax Species in Vietnam. Biosc.Biotech.Res.Comm. 2021;14(2). Available from: <a href=”https://bit.ly/3bMHFlB“>https://bit.ly/3bMHFlB</a>
Copyright © Nguyen et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Ginseng is the common name of the species in the Panax genus Araliaceae. This plant has been used as medicine for a long time as its outstanding medicinal properties include stimulating metabolism in the body, regulating blood sugar in the body, and protecting the activity of the central nervous system, counteract the formation and inhibit the growth of tumors, increase the body’s resistance to diseases (Ali et al., 2012; Peng et al., 2012; Lee et al., 2015).
High medicinal activity of ginseng is due to the plant’s high ginsennoside content, it is also rich in other valuable ingredients such as anti-oxidant compounds, peptides, polysaccharides, and vitamins (Lee et al., 1995; Hong et al., 2012). So far, at least 17 Panax species have been reported worldwide (Zhang et al., 2000) and medicinal compounds are variable from species to species (Harkey et al., 2001; Liu et al., 2016, Hyeonah et al 2021 ).
Currently, ginseng identification and classification is mainly based on morphological characteristic since it is straightforward to perform and feasible to carry out on the field at a low cost (Proctor et al., 2011; Hong et al., 2012). Nevertheless, significant limitations of this method have been addressed such as low number, complex inheritance pattern, and vulnerable to changes in the environment (Ahmedand and Mohamed, 2014). Consequently, ginseng authentication and cultivar identification based on this method are unreliable.
With the rapid development in molecular biology, DNA-based markers have been used intensively for characterizing the genetic relationships among plant species due to their fast, specific, reliable, and sensitive features (Shim et al., 2003; Serrone et al., 2006; Kim et al., 2016). Recently, the DNA barcode has been used intensively for classifying at the species level (CBOL Plant Working Group, 2009). This marker type has been applied to discriminate at species level for different organism such as plants, animal, fungi and bacteria (Hollingsworth et al., 2011; Barcaccia et al., 2016; Hu et al., 2019).
Among several DNA barcode loci, Internal Transcribed Spacer (ITS) region has been proposed as a standard region for plant identification because the ITS region is highly conserved among interspecies but variable between interspecies and is easy to amplify even from limited DNA quantity (Chase et al., 2007; Giudicelli et al., 2015). Numerous researchers reported that discrimination power of ITS is higher in the comparison to other plastid markers (Muellner et al., 2011; Yang et al., 2012; Zhang et al., 2014).So far, several studies have exploited the ITS-based DNA barcode in the investigating the genetic composition and developing protocols to differentiate species in Panax genus. In 2011, a study in India showed the effectiveness of ITS in clustering analysis of Panax assamicus populations according to geographic locations (Panday and Ali, 2011).
In Saudi Arabia, Ali and colleagues applied ITS2 sequence to control ginseng-derived products (Ali et al., 2012). Bang and colleagues used ITS to differentiating and authenticating Panax species in Korea (Bang et al., 2015). More recently, this barcode region was also applied to characterize genetic diversity of 24 ginseng samples collected at Lai Chau province of Vietnam (Pham et al., 2018). The present study describes the effectiveness of ITS-based DNA barcode in classifying 23 Panax samples belong to different species collected in different geographical locations in Vietnam. The obtained data could be applied for classification, identification, and authentication activities in Panax plants of Vietnam.
MATERIAL AND METHODS
Sample collection:Total of 23 Panax samples were collected from North to South of Vietnam, samples were collected from different sources such as Panax traders, research institutes, universities, and companies in different locations (Figure 1 and Table 1). Collected samples were then store in cool and dry condition and targeted for DNA extraction.
Figure 1: Map showing locations for collecting Panax samples.
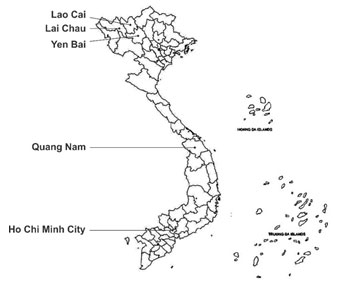
Table 1. List of collected Panax samples used in this study.
| No. | Sample ID | Local name | Collection location |
| 1 | TTB1 | Tam that bac | Quang Nam province |
| 2 | TTB2 | Tam that bac | Quang Nam province |
| 3 | TTH1 | Tam that hoang | Lao Cai province |
| 4 | TTH2 | Tam that hoang | Yen Bai province |
| 5 | TT1 | Tam that | Quang Nam province |
| 6 | TT2 | Tam that | Quang Nam province |
| 7 | TT3 | Tam that | Quang Nam province |
| 8 | TT4 | Tam that | Quang Nam province |
| 9 | TT5 | Tam that | Quang Nam province |
| 10 | TT6 | Tam that | Quang Nam province |
| 11 | TT7 | Tam that | Quang Nam province |
| 12 | TT8 | Tam that | Quang Nam province |
| 13 | TT9 | Tam that | Quang Nam province |
| 14 | TT10 | Tam that | Quang Nam province |
| 15 | SLC1 | Sam Lai Chau | Lai Chau province |
| 16 | SLC2 | Sam Lai Chau | Lai Chau province |
| 17 | SNL1 | Sam Ngoc Linh | Ho Chi Minh city |
| 18 | SNL2 | Sam Ngoc Linh | Quang Nam province |
| 19 | SNL3 | Sam Ngoc Linh | Quang Nam province |
| 20 | SNL4 | Sam Ngoc Linh | Quang Nam province |
| 21 | SVD1 | Sam vu diep | Yen Bai province |
| 22 | SVD2 | Sam vu diep | Lao Cai province |
| 23 | SVD3 | Sam vu diep | Yen Bai province |
DNA extraction:DNA from Panax samples extracted by cetyltrimethyl ammonium bromide (CTAB) method followed Li et al. (2013). After extraction, DNA quality was examined by electrophoresis on 1% agarose then spectrophotometer (Optima SP 3000 nano UV-VIS, Japan) was used to determine DNA concentrations. The DNA samples were then kept at -20 °C freezer until use for PCR reactions.
PCR reactions and DNA sequencing:The composition of PCR reactions to amplify ITS region as follows: 7.5 μL 2X Mytaq Red Mix (Bioline, UK), 20 ng DNA, 0.2 μM of each primer (C26A 5’ AGGAGAAGTCGTAACAAG3’ and N-nc18S10 5’ GTTTCTTTTCCTCCGCT 3’) (Wen and Zimmer, 1996), and PCR water for a final volume of 15 μl. The PCR cocktails were run in thermal cyclers SureCycler 8800 Thermal Cycler (Agilent, USA) with following conditions: initial denaturation at 94 °C for 2 minutes; then repeated by 35 cycles of 30 seconds at 94 °C, 30 seconds at 55 °C, 50 seconds at 72 °C, and finally one minute at 72 °C to complete the reaction. The PCR products were visualized on gel electrophoresed and use 1 kb ladder (Bioline, UK) to determine amplification length.
Correct PCR products were sequenced by Sanger methods at Nam Khoa Company (Ho Chi Minh City, Vietnam). Each sample were sequenced for both sense and antisense directions. Nucleotide sequences of both DNA strands were obtained and compared the forward and reverse sequence to ensure accuracy. The finally assembled sequences were submitted to NCBI GenBank to obtained accession number (Table 2) and then used for following analysis.
Data analysis: The homology identification of each sequence were implemented by using Basic Local Alignment Tools (BLAST) function in NCBI (National Center for Biotechnology Information, USA) database. To evaluate the capacity of ITS based DNA barcode in distinguish Panax accessions from Vietnam and abroad, additional ten ITS sequences of related Panax species retrieved from Genbank. All sequences were aligned using the Clustal method in Molecular Evolutionary Genetics Analysis (MEGA) 6 software (https://www.megasoftware.net).
Evolutionary trees were constructed based on two methods consisting of Maximum Likelihood (ML) and Neighbour Joining (NJ) since they represent for discrete character methods and distance methods, respectively. For satisfactory reliability in phylogenetic construction, bootstrap value was set at 1000 replicates and bootstrap support was classified as Kress et al. (2002).
RESULTS AND DISCUSSION
PCR and DNA sequence: Although three are few study reported the difficulty in amplifying and sequencing ITS region (Gonzalez et al., 2009; Wang et al., 2016), all PCR and sequencing reactions in this study were performed successfully. The sequences length is 693 bp on average, varying from 631 bp to 707 bp. The similarity of obtained sequences to homologous sequences in NCBI Genbank ranges from 99.52 to 100%. The Latin names of collected samples were identified and presented in Table 2.
Table 2. Genbank accession numbers of samples and corresponding Latin names.
| No. | Sample ID | Length (bp) | Accession number | Latin name | Percent identity (%) |
| 1 | TTB2 | 704 | MZ149934 | Panax notoginseng | 100 |
| 2 | TTB1 | 707 | MZ149935 | Panax notoginseng | 99.86 |
| 3 | TTH1 | 703 | MZ149936 | Panax stipuleanatus | 99.72 |
| 4 | TTH2 | 631 | MZ149937 | Panax stipuleanatus | 99.68 |
| 5 | TT1 | 705 | MZ149938 | Panax japonicus var. bipinnatifidus | 99.43 |
| 6 | TT2 | 702 | MZ149939 | Panax japonicus var. bipinnatifidus | 99.57 |
| 7 | TT3 | 702 | MZ149940 | Panax japonicus var. bipinnatifidus | 99.57 |
| 8 | TT4 | 702 | MZ149941 | Panax japonicus var. bipinnatifidus | 99.57 |
| 9 | TT5 | 702 | MZ149942 | Panax japonicus var. bipinnatifidus | 99.57 |
| 10 | TT6 | 702 | MZ149943 | Panax japonicus var. bipinnatifidus | 99.57 |
| 11 | TT7 | 701 | MZ149944 | Panax japonicus var. bipinnatifidus | 99.43 |
| 12 | TT8 | 702 | MZ149945 | Panax japonicus var. bipinnatifidus | 99.57 |
| 13 | TT9 | 702 | MZ149946 | Panax japonicus var. bipinnatifidus | 99.57 |
| 14 | TT10 | 702 | MZ149947 | Panax japonicus var. bipinnatifidus | 99.57 |
| 15 | SLC1 | 700 | MZ149948 | Panax vietnamensis var. fuscidiscus | 99.86 |
| 16 | SLC2 | 630 | MZ149949 | Panax vietnamensis var. fuscidiscus | 99.52 |
| 17 | SNL1 | 704 | MZ149950 | Panax vietnamensis | 100 |
| 18 | SNL2 | 701 | MZ149950 | Panax vietnamensis | 99.71 |
| 19 | SNL3 | 702 | MZ149951 | Panax vietnamensis | 99.72 |
| 20 | SNL4 | 705 | MZ149952 | Panax vietnamensis | 99.86 |
| 21 | SVD1 | 698 | MZ149953 | Panax stipuleanatus | 100 |
| 22 | SVD2 | 703 | MZ149954 | Panax stipuleanatus | 99.72 |
| 23 | SVD3 | 642 | MZ149955 | Panax stipuleanatus | 99.53 |
After identification, 23 samples were found belonging to four species consisting of Panax notoginseng, Panax stipuleanatus, Panax japonicus var bipinnatifidus, Panax vietnamensis var. fuscidiscus, and Panax vietnamensis Thus, two different plant groups in Vietnam local names consisting of “Tam that hoang” including TTH1 and TTG2 samples and “sam vu diep” including SVD1, SVD2 and SVD3 samples were identified as Panax stipuleanatus. This confirm the low accuracy in traditional method for Panax classification due to several uncontrollable factors such as changes of environmental conditions and developmental stages (Ali et al., 2012).
Sequence alignment and Phylogenetic analysis: Together with 23 obtained sequences from this study, additional 10 ITS sequences from different species in Panax genus were downloaded from NCBI Genbank and combined for sequence alignment and the alignment summary is presented in Table 3. Totally, up to three 33 variation sites were detected within analyzed sequences. Previously, several research groups have employed the nucleotide variations to develop molecular makers for plant identification such as in Taxus (Liu et al., 2011) or onion (Ipek et al., 2014). Thus, these variations are valuable for developing specific markers or diagnosis KIT for discriminating different Panax species.
Table 3. Variable sites in 33 ITS sequences from different Panax species.
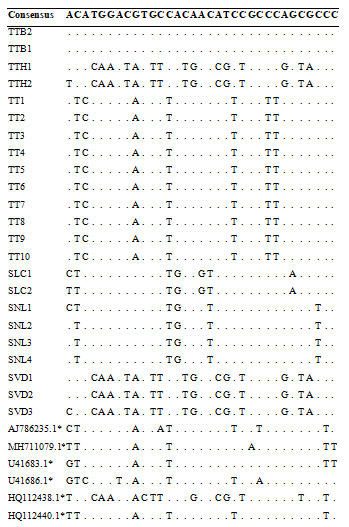
(*: sequences were retrieved from NCBI Genbank)
The genetic distances among 23 Panax samples were calculated based on Kimura-2 parameter (K2P) method of MEGA 6 software. The lowest genetic distance is 0.000 indicating the sequence similarity among samples, while the highest is 0.052 this result showing the close genetic relationship among these accessions when using ITS regions. The phylogenetic trees constructed by either neighbor-joining (NJ) or Maximum Likelihood methods are shown in Figure 2 and Figure 3, respectively.
Figure 2: Neighbor-Joining tree with 1000 bootstrap replicated based on ITS sequences (Star symbols indicating sequences retrieved from NCBI Genbank and the availability of common names are shown in parentheses).
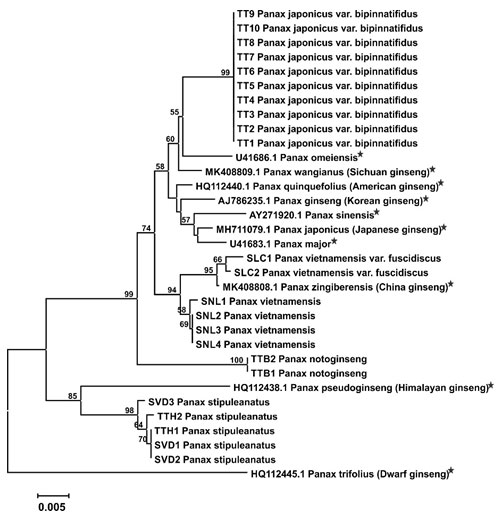
Figure 3: Maximum Likelihood tree with 1000 bootstrap replicated based on ITS sequences (Star symbols indicating sequences retrieved from NCBI Genbank and the availability of common names are shown in parentheses).
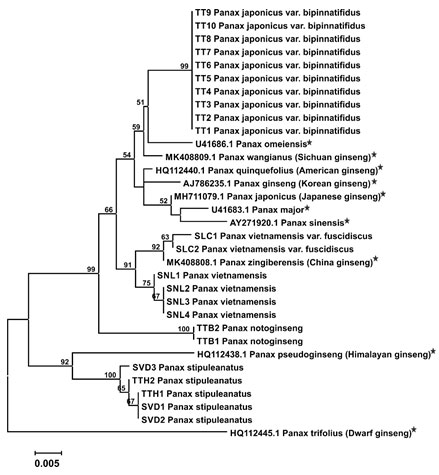
In general, the results show the high similarity between Neighbor-Joining and Maximum Likelihood methods. Panax samples collected from Vietnam in this study is clustered into several groups corresponding to its identified species using NCBI BLAST. Interestingly, Vietnam ginsengs are also separated from different ginseng species collected from different countries. The grouping of Panax species in obtained phylogenetic trees of this study is slightly different with previous study of Yang and colleagues in Korea in which they reported the closer clustering of P. notogingsen and P. pseudoginseng as well as P. omeiensis and P. zingiberensis (Yang et al., 2001).
CONCLUSION
The obtained data from this study confirms the effectiveness of using ITS marker to classify Panax samples in Vietnam. Furthermore, this marker also shows high discrimination power to distinguish Panax species from Vietnam and from abroad. Future study should focus on develop specific primer pairs or diagnosis KIT to serve for identification and authentication activities of Panax plants.
Ethics approval and consent to participate: Not applicable.
Conflict of Interests: The author has no conflict of interest.
Funding statement: This work was sponsored and funded by Ho Chi Minh City University of Food Industry under Contract No. 53/HD-DCT.
ACKNOWLEDGEMENTS
The authors would like to thank Ho Chi Minh University of Food Industry for providing research fund and laboratory facility.
REFERENCES
Ahmedand THM, Mohamed ZMA (2014). Genetic diversity of mango (Mangifera indica L.) cultivars in Shendi Area. Extensive journal of applied sciences 3 (6): 219-224.
Ali MA, Al-Hemaid FM, Lee J et al. (2012) Assessing nrDNA ITS2 sequence based molecular signature of ginseng for potential use in quality control of drug. African Journal of Pharmacy and Pharmacology Vol 6 No 39 Pages 2775-2781. DOI: 10.5897/AJPP12.384
Bang KH, Kim YC, Lim JY et al (2015) Internal Transcribed Spacer Barcoding DNA Region Coupled with High Resolution Melting Analysis for Authentication of Panax Species. Korean Journal of Medicinal Crop Science Vol 23 No 6 Pages 439−445. http://dx.doi.org/10.7783/KJMCS.2015.23.6.439
Barcaccia G, Lucchine M, Cassandro M (2016). DNA Barcoding as a Molecular Tool to Track Down Mislabeling and Food Piracy. Diversity Vol 8 No 2 doi:10.3390/d8010002.
CBOL Plant Working Group et al (2009). A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America Vol 106: 12794-12797.
Chase MW, Cowan RS, Hollingsworth PM et al (2007). A proposal for a standardized protocol to barcode all land plants. Taxon Vol 56 Pages 295–299.
Giudicelli GC, Mader G, Freitas LBD (2015). Efficiency of ITS sequences for DNA barcoding in Passiflora (Passifloraceae). International Journal of Molecular Sciences Vol 16 Pages7289–7303.
Gonzalez MA, Baraloto C, Engel J et al (2009). Identification of Amazonian Trees with DNA Barcodes. PLoS ONE Vol 4 No 10: e7483. https://doi.org/10.1371/journal.pone.0007483.
Harkey MR, Henderson GL, Gershwin ME et al (2001). Variability in commercial ginseng products: an analysis of 25 preparations. The American Journal of Clinical Nutrition Vol 73 Pages 1101-1106.
Hollingsworth PM, Graham SW, Little DP (2011). Choosing and using a plant DNA barcode. Plus ONE Vol 6 No 5: e19254 doi:10.1371/journal.pone.0019254
Hong HD, Cho CW, Kim YC et al (2012). Morphological Characteristics of Korean Dried Ginseng Products. Journal of Ginseng Research Vol 36 No 3 Pages 314-321. DOI: 10.5142/jgr.2012.36.3.314.
Hu SJ, Hu HY, Gao H et al. (2019). DNA barcode and rapid identification of the precious herb Herba Anoectochili. Chinese journal of Natural medicines 17(10): 0738-0745.
Hyeonah, Shim, Nomar Espinosa Waminal, Hyun Hee Kim & Tae-Jin Yang (2021) Dynamic evolution of Panax species Genes & Genomics volume 43, pages 209–215
Ipek M, Ipek A, SimonWP (2014). Testing the utility of matK and ITS DNA regions for discrimination of Allium species. Turkish Journal of Botany Vol 28 Pages 203-212.
Kim JH, Kim MK, Wang H et al (2016). Discrimination of Korean ginseng (Panax ginseng Meyer) cultivar Chunpoong and American ginseng (Panax quinquefolius) using the auxin repressed protein gene. Journal of Ginseng Research Vol 40 No 4 Pages 395-399. https://doi.org/10.1016/j.jgr.2015.12.002.
Kress WJ, Wurdack KJ, Zimmer EA et al (2002) Use of DNA barcodes to identify flowering plants, Proceedings of the National Academy of Sciences of the United States of America Vol 102 No 23 Pages 8369–8374.
Lee HS, Kim SW, Lee SW et al (1995). Agrobacterium mediated transformation of ginseng (P. ginseng) and mitotic stability of the inserted beta-glucuronidase gene in regenerates from isolated protoplasts. Plant Cell Reports Vol 14 No 9 Pages 545-549. doi: 10.1007/BF00231935
Lee SM, Bae BS, Park HW et al (2015) Characterization of Korean Red Ginseng (Panax ginseng Meyer): History, preparation method, and chemical composition. Journal of Ginseng Research Vol 39 No 4 Pages 384-391. https://doi.org/10.1016/j.jgr.2015.04.009
Li J, Wang S, Jing Y et al (2013). A modified CTAB protocol for plant DNA extraction. Chinese Bull Bot Vol 48 Pages 1-7.
Liu J, Moller M, Ga LM et al (2011) DNA barcoding for the discrimination of Eurasian Yews (Taxus L., Taxaceae) and the discovery of cryptic species. Molecular Ecology Resources Vol 11Pages 89-100.
Liu J, Liu Y, Zhao L et al (2016). Profiling of ginsenosides in the two medicinal Panax herbs based on ultra-performance liquid chromatography-electrospray ionization–mass spectrometry. Springer Plus Vol 5, 1770. https://doi.org/10.1186/s40064-016-3427-3
Muellner AN, Schaefer H, Lahaye R (2011). Evaluation of candidate DNA barcoding loci for economically important timber species of the Mahogany family (Meliaceae). Molecular Ecology Resources Vol 11 Pages 450–60.
Panday AK, Ali MA (2011). Intraspecific variation in Panax assamicus Ban. Populations based on internal transcribed spacer (ITS) sequences of nrDNA. Indian Journal of Biotechnology Vol 11 Pages 30-38.
Peng D, Wang H, Qu C et al (2012) Ginsenoside Re: Its chemistry, metabolism and pharmacokinetics. Chinese Medicine Vol 7 No 2. https://doi.org/10.1186/1749-8546-7-2
Pham QT, Nguyen MD, Khuong TB et al (2018) Genetic diversity of several gingseng plants collected in Lai Chau. Vietnam Journal of Science, Technology and Engineering Vol 60 No 2 Pages 27-31.
Proctor JTA, Sullivan AJ, Rupasinghe HPV et al (2011) Morphological and Ginsenoside Differences among North American Ginseng Leaves. Journal of Ginseng Research Vol 35 No 2 Pages 155-161. DOI: 10.5142/jgr.2011.35.2.161
Serrone PD, Attorri L, Gallinella B et al (2006) Molecular Identification of Panax ginseng C.A. Meyer in Ginseng Commercial Products. Natural Product Communications Vol 1 No 12 Pages 1137-1140. DOI: 10.1177/1934578X0600101213
Shim YH, Choi JH, Park CD et al (2003) Molecular differentiation of Panax species by RAPD analysis. Archives of Pharmacal Research Vol 26 No 8 Pages 601-605.
Wang A, Gopurenko D, Wu H et al (2016) Evaluation of six candidate DNA barcode loci for identification of five important invasive grasses in eastern Australia. PloS One Vol 12 No 4: e0175338
Wen J, Zimmer EA (1996) Phylogeny and Biogeography of Panax L. (the Ginseng genus, Araliaceae): Inferences from ITS sequences of Nuclear Ribosome DNA. Molecular Phylogenetics and Evolution Vol 6 No 2 Pages 167-177.
Yang DC, Yang KJ, Yoon ES (2001) Comparison of ITS (Internal Transcribed Spacer) and 5.8S rDNA sequences among varieties and cultivars in Panax ginseng. Journal of Photoscience Vol 8 No 2 Pages 55-60.
YangJB, Wang YP, Möller M et al (2012) Applying plant DNA barcodes to identify species of Parnassia (Parnacciaceae). Molecular Ecology Resources Vol 12 Pages 267–75.
Zhang D, Jiang B, Duan L et al (2016) Internal transcribed spacer (ITS), an ideal DNA barcode for species discrimination in Crawfurdia wall (Gentianaceae). African Journal of Traditional, Complementary and Alternative Medicines Vol 13 No 6 Pages 1010-1106.
Zhang H, Abid S, Ahn JC et al (2020) Characteristics of Panax ginseng cultivars in Korea and China. Molecules Vol 25 No11 2635. doi.org/10.3390/molecules25112635


