1State Key Laboratory of Crop Genetics and Germplasm Enhancement, College of Horticulture, Nanjing Agricultural University, Nanjing, Jiangsu, 210095, P.R. China
2Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement, Jiangsu Academy of Agricultural Sciences, Nanjing, Jiangsu, 210014, P.R. China.
Corresponding author Email: changweizh@njau.edu.cn
Article Publishing History
Received: 17/04/2019
Accepted After Revision: 18/06/2019
The SAND domain protein ULTRAPETALA1 (ULT1) and its paralog ULT2 define a small family of closely related plant proteins, both of them physically associated to form heterodimers of trxG (Trithorax group) factors. Although trxG gene ULT1 has well-defined roles in controlling gynoecium patterning and cell fate decisions of plants, relatively little is known about the specific functions of ULT2. In this study, bioinformation analysis revealed that the BrULT2 gene of Brassica rapa encodes a protein of molecular mass of 26.37 KDa, an isoelectric point of 7.9. qRT-PCR was performed to detect the expression profiles of the BrULT2 gene of Chinese cabbage, showing that the transcription level of BrULT2 gene was significantly down-regulated during the vegetative stage. We silenced the trxG family gene BrULT2 with optimized TYMV induced gene silencing, the TYMV-derived vector pTY was recombinated to infect Brassica rapa. After twenty days of inoculation, the infected plants showed abnormal phenotypes: leaf rolling, smaller and increased number leaves, thin stem, branch outgrowth. We detected that the expression level of BrULT2 gene of infected plants was down-regulated meanwhile the transcriptional level of BrULT1 gene was up-regulated.
BrULT1, BrULT2, Chinese cabbage, leaf shape, VIGS
Li R, Song L, Yu J, Hou X, Zhang C. TYMV Induced Gene Silencing of TrxG Family Gene BrULT2 Affects the Leaf Shape of Chinese Cabbage. Biosc.Biotech.Res.Comm. 2019;12(2).
Li R, Song L, Yu J, Hou X, Zhang C. TYMV Induced Gene Silencing of TrxG Family Gene BrULT2 Affects the Leaf Shape of Chinese Cabbage. Biosc.Biotech.Res.Comm. 2019;12(2). Available from: https://bit.ly/2WrPIyR
Copyright © Li et al, This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
The epigenetic factors Trithorax group (trxG) and Polycomb group (PcG) proteins act as antagonistic regulators to maintain gene in transcriptional repressed and active states through their histone lysine methyltransferase (HKMT) activity during plant developmental processes (Bernadett and Jürg, 2006, Maria et al., 2015). Historically, the two major and best-characterized PcG proteins are Polycomb repressive complex 1 (PRC1) and PRC2 (Zheng and Chen, 2011), several proteins with trxG-like functions also have been identified in plants: ARABIDOPSIS HOMOLOG OF TRITHORAX1 (Alvarez-Venegas et al., 2003), ATX2 (Saleh et al., 2008), ATXR3/SDG2 (Alexandre et al., 2010, Lin et al., 2010), ATXR7/SDG25 (Berr et al., 2009, Tamada et al., 2009), and the SAND-domain DNA binding proteins ULTRAPETALA1 (ULT1) and ULT2 (Carles et al., 2005). Previous reports have demonstrated that the Arabidopsis trxG gene ULT1 can switch the key floral homeotic gene AGAMOUS (AG) locus from repressed state to active state through restricting the deposition of H3K27 methylation, which was mediated by CURLY LEAF (CLF) containing the PRC2 complexes (Carles et al., 2009, Goodrich et al., 1997). The ULT1 gene also acts antagonistically to KAN1 to pattern adaxial-abaxial polarity of gynoecium and rosette leaves (Pires et al., 2014, Pires et al., 2015). The trxG gene ULT2 have a very similar amino acid sequence and mRNA expression pattern with ULT1 (Carles et al., 2005), but there are little researchs focus on the specific biological function of ULT2 gene. To gain insight into the molecular mechanism of ULT2 activity in plant, the optimized VIGS technology was performed.
Virus-induced gene silencing (VIGS) is an RNA mediated specific degradation mechanism based on highly conserved nucleic acid levels. By using the viral vector carrying the cDNA fragment of the target gene to infect plant, VIGS can specifically induce the sequence degradation or methylation of the homologous gene along with the replication and transcription of the virus, that will generate changes in plant phenotype or physiological indicators in favour of identifing the functional gene (Burch-Smith et al., 2010, Vaghchhipawala et al., 2011). In 1995, Kumagai constructed a VIGS vector based on the tobacco mosaic virus (TMV) for the first time, then the VIGS was successfully performed to silencing the expression of phytoene desaturase gene PDS in tobacco (Nicotiana benthamiana). Nowadays, VIGS system has been successfully established not only on model plants such as Arabidopsis (Manhães et al., 2015), Nicotiana benthamiana and Solanum lycopersicum (Velásquez et al., 2009), but also on wheat (Bennypaul et al., 2012), cotton (Ye et al., 2014), rice (Kant et al., 2015). Turnip yellow mosaic virus (TYMV) is a higher plant virus with a positive RNA strand, which can infect many cruciferous plants (Martínezherrera et al., 1994). Recently improved VIGS system based on TYMV-derived vector has been exploited, which has been successfully performed by gun powder with synthesized 80-nt fragment identical to target gene (Yu et al., 2018).
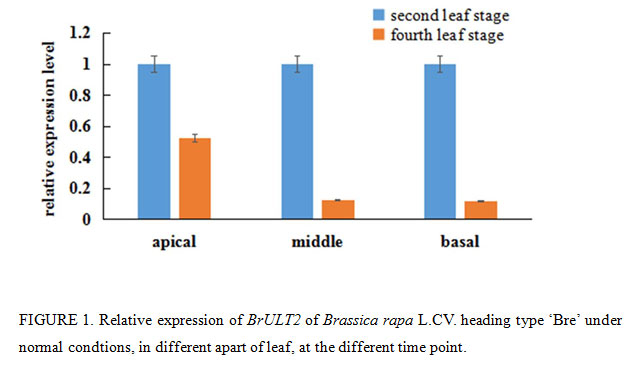 |
Figure 1: Relative expression of BrULT2 of Brassica rapa L.CV. heading type ‘Bre’ under normal condtions, in different apart of leaf, at the different time point. |
Previous study have indicated that the trxG genes ULT1 and ULT2 have overlapping roles in regualting shoot and floral stem cell accumulation during the reproductive period, however, they have not been sufficiently characterized during the vegetative growth stage. Here, we characterized the role of BrULT2 gene of Brassica rapa, optimized VIGS system was deployed to silenced the BrULT2 gene, qRT-PCR was performed to decideted the expression level of BrULT gene of innoculated and wild type Chinese cabbage, and the morphology of BrULT2-silenced transgenic plants was investigated. The collective results of our study will provide some basic information related to the meshanism of leaf growth of Chinese cabbage.
Materials and Methods
Plant Materials and VIGS Assay
Brassica rapa L.CV. heading type ‘Bre’ and non-heading type ‘49 caixin’ were grown in soil (1:1:1 mixture of perlite : vermiculite : topsoil) and placed in artificial climate room at 22℃ with 16h/8h light/dark photoperiod cycle, 50% relative humidity. When the cotyledons and the first 3 true leaves have emerged, the empty vector pTY-S and recombined vector pTY-PDS and pTY-BrUL T2 were transformed into plants by applying 10μL 2mg cm -3 plasmid to leaves which was injured advanced by rubbing with quartz sand. Three weeks post-inoculation, leaf samples were collected directly from the control plant and BrULT2-silenced plant, it was frozen with liquid nitrogen and stored at -80℃ until used. All experiment were conducted with three biological replicates.
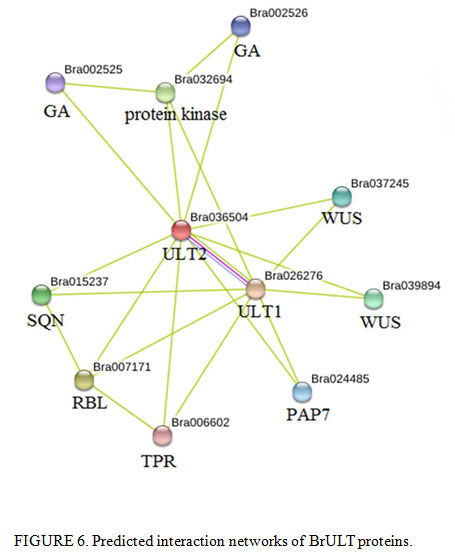 |
Figure 6: Predicted interaction networks of BrULT proteins |
Plasmid Construction
The sequence of BrULT2 gene was downloaded from the Brassica database (http://brassicadb.org/brad/), total RNA was extracted from the tissue of heading type ‘Bre’. The open reading frame (ORF) of BrULT2 gene was amplified by PCR, using first-strand cDNA as template and gene-specific primers which are listed in Table 1. The PCR conditions were: 95 ℃ for 5 min, followed by 35 cycles at 95 ℃ for 30 s, 55 ℃ for 30 s and 72 ℃ for 1 min, with a final extension at 72 ℃ for 10 min. The PCR products were detected by 1.2% agarose gel electrophoresis and recovered with AxyPrep DNA gel extraction kit (AxyPrep, China), then they were cloned into the pMD19-T vector (TaKaRa, Japan) and verified by sequencing. 40-nt fragment identical to BrULT2 gene was selected for plasmid construction (5’-TTAGGGTTTTCTCAGATGGAGACCTCCAAATCACTTGCCA-3’), specific method has been described in Yu et al (2018). The fragment was self-hybridized and the empty vector pTY-S was digested with SnaBI, then the 80-nt inverted-repeat fragment of BrULT2 was inserted into the linearized vector pTY-S at 3:1 ratio. Afterwards, the recombined vector BrULT2-pTY, pTY-PDS and the empty vector pTY-S were transformed into Escherichia coli strain DH5α cells and subjected to sequencing. The empty vector and recombinant vector were isolated using maxiprep-quality plasmid (Tiangen, China) and concentrated to 2mg cm -3 for VIGS assay.
Bioinformatics Analysis of ULT family genes and Conserved Motif Analysis
A phylogenetic tree of the ULT family genes was generated using the MEGA (version 6.0) program with the neighbor-joining method based on multiple alignments of their protein amino acid sequences, the internal branch support was estimated using 1,000 bootstrap replicates. The ULT genes of Arabidopsis thaliana were obtained from the TAIR database (http://www.arabidopsis.org/), the ZmULT1 gene was searched from NCBI database (https://www.ncbi.nlm.nih.gov), and the BrULT gene of Brassica rapa and BolULT gene of Brassica oleracea were downloaded from Brassica database (http://brassicadb.org/brad/), all accession numbers are listed in Table 2. Multiple sequences alignment of BrULT1 protein and BrULT2 protein was performed by using DNAMAN software (version 5.0). The EMBOSS Pepstats software was used to calculate statistics of protein properties. Conserved motif analysis used the Multiple Expectation Maximization for Motif Elicitation (MEME) (http://meme.nbcr.net/meme/) search tool. The protein secondary structure was predicted by predictprotein (https://www.predictprotein.org/), gene interaction model was analysised by SMART (Main page http://smart.embl-heidelberg.de/).
Table 1: Sequences of the specific Primers used in gene cloning.
| gene name | Sense Primer | Anti-sense Primer |
| BrULT2 | 5’-ATGGAGAGAGAATGCGGGTCGA-3’ | 5’-TCAAATGGGTTTGGTGTTGG-3’ |
Isolation of RNA and cDNA Synthesis
Total RNA was extracted from the leaves of BrULT2-pTY and pTY-S inoculated plant by using the RNAeasymini kit (TIANGEN, China). Subsequently, the first-strand cDNA was synthesized with 1μg of total RNA according to the instructions of the manufacturer’s PrimeScript TM1 st Strand cDNA Synthesis Kit (TaKaRa, Japan).
Gene Expression Analysis by qRT-PCR
The Gene-specific primers were designed based on the gene sequences of BrULT1 and BrULT2 using the Becon Designer v. 7.9, that are listed in Table 3. The analysis was performed on 7500 Fast Real-Time PCR System (Applied Biosystems, USA) following the instruction manual for the SYBR Green (TaKaRa, Japan). The thermocycling conditions were as follows: initial polymerase activation at 94 °C for 30s; followed by 45 cycles at 94 °C for 10s , at 60 °C for 30s; and finally a melting curve was performed (60 cycles at 65 ℃for 10 s). Each reaction was performed in triplicate. The Actin gene (Bra028615) was used as endogenous control (Dheda et al. 2004), and the relative expression levels of BrULT1 and BrULT2 gene were calculated using the 2-∆∆CT method as described by Schmittgen and Livak (2008).
Results and Discussion
Expression of BrULT2 during the Vegetative Stage of Chinese Cabbage
In Arabidopsis thaliana, the ULT1 and ULT2 gene are expressed coordinately in shoot and floral meristems, developing stamens, carpels and ovules, the ULT1 transcripts could also be detected in vegetative meristems and leaf primordia during the vegetative stage, while the ULT2 expression is specific to the reproductive developmental stage, which can be detected only in inflorescences, pollen and siliques (Carles et al., 2005). In this study, qRT-PCR was performed to determine the distribution of BrULT2 mRNA transcripts in Brassica rapa L.CV. heading type ‘Bre’, the specific primer pairs are shown in Table 3. The analysis results showed that the BrULT2 mRNA transcripts can be detected in the wild type leaves during vegetative stage, and it decreased significantly from the second leaf stage to fourth leaf stage with more than 80% drop at the middle and basal region of the leaf, 50% drop at apical portion (Fig. 1).
Sequences Analysis of BrULT genes
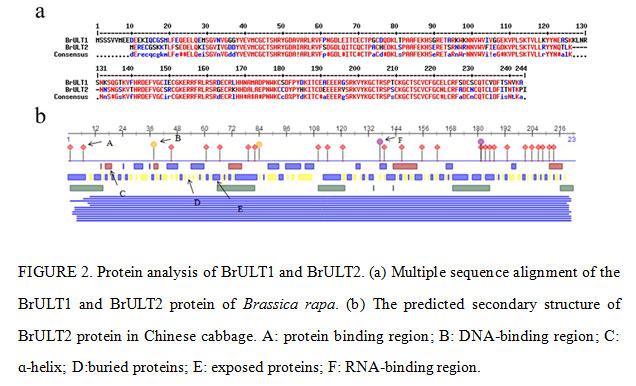
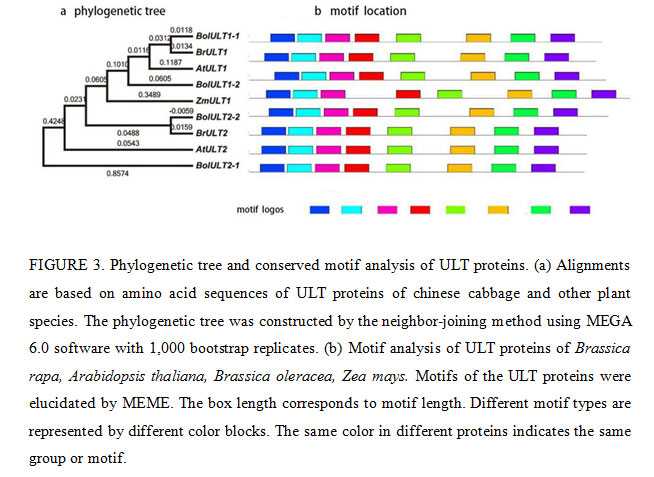
Bioinformatics analysis of ULT family genes showed that the predicted molecular weights (MW) of the ULT1 and ULT2 proteins ranged from 27.39 kDa to 8.98 kDa, and the theoretical isoelectric points (pIs) varied from 7.90 to 6.83 (Table 2). The cDNA sequence of BrULT2 gene was 639bp in length, the calculated molecular mass of the protein encoded by BrULT2 was 26.37 kDa with theoretical pI of 7.90. Multiple sequence alignment showed that the sequences of BrULT1 protein and BrULT2 protein were basically the same (Fig. 2a). To better understand the similarities and differences of ULT family genes between Brassica rapa and other plants, an unrooted phylogenetic tree was generated, the analysis result showed that the ULT family genes were highly conserved among the development of Arabidopsis thaliana, Zea mays, Brassica rapa, Brassica oleracea (Fig. 3a), and the BrULT2 (Brassica rapa) and BolULT2-2 (Brassica oleracea) clustered on the same branch. A total of 8 motifs were identified by analysis amino acid conserved domains of the ULT genes, showing that the conserved domain structure and positions were homogeneous (Fig. 3b). The prediction of secondary structure of BrULT2 gene-encoding protein showed that BrULT2 protein has multiple RNA and protein binding sites (Fig. 2b), which may facilitate its interaction with multiple transcription factors and contribute to its close regulation of a variety of growth and development-related genes.
Table 2: ULT family genes identified in Brassica rapa, Arabidopsis thaliana, Brassica oleracea, Zea mays and the sequence characteristics.
| Gene name | Locus ID | Chromosome | Start | Stop | Isoelectric
point |
Molecular weight(KDa) |
| BrULT1 | Bra026276 | A01 | 10038908 | 10040319 | 7.0849 | 27397.08 |
| BolULT1-1 | Bol020015 | Scaffold000127 | 1378942 | 1380353 | 6.8382 | 26993.52 |
| BolULT1-2 | Bol042381 | C06 | 44651418 | 44652931 | 7.7838 | 26673.57 |
| AtULT1 | AT4G28190 | Chr4 | 13985178 | 13987442 | 7.2871 | 26744.24 |
| ZmULT1 | NC_024466.2 | Chr8 | 177291861 | 177295258 | 7.8795 | 26486.92 |
| BrULT2 | Bra036504 | A07 | 33825 | 34786 | 7.9079 | 26375.91 |
| BolULT2-1 | Bol001467 | Scaffold000442 | 29537268 | 29540037 | 6.9431 | 8982.30 |
| BolULT2-2 | Bol001263 | Scaffold000457 | 120188 | 121156 | 7.6502 | 26378.92 |
| AtULT2 | AT2G20825 | Chr2 | 8966867 | 8969756 | 7.5056 | 26097.69 |
VIGS of BrULT2 was Successfully Performed in Brassica rapa
Comparing with genetic research methods such as transgene, gene knockout, and antisense inhibition, VIGS hardly require genetic transformation and mutant acquisition with a short research cycle, according to its outstanding merits: low cost, simple to operate, which has become one of the most attractive technological approaches in functional genomics research (Becker and Lange 2010). In this study, the optimized VIGS was performed to gain insight into the specific function of BrULT2 gene.
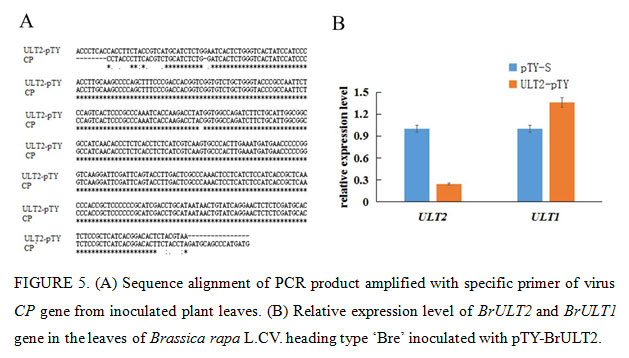
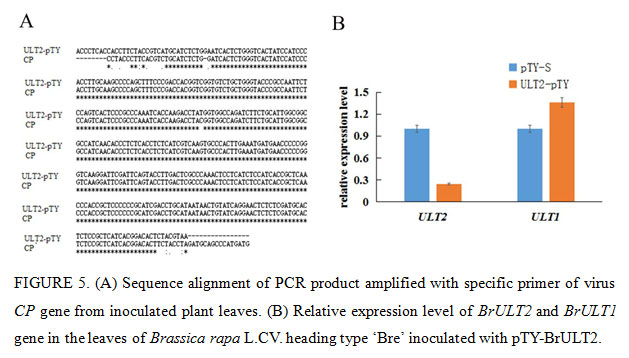
The Phytoene desaturase (PDS) protein is a key enzyme in the carotenoid synthesis pathway, plants will lose the photoprotective effect of carotenoids and thus exhibit a whitening effect when the expression of PDS was blocked (Velásquez et al., 2009). In present study, the specific 40np of BrULT2 and BrPDS CDS of the Brassica rapa was selected to construct intronic hair pin silencing vector to targeting the native BrULT2 and BrPDS transcripts, the antisense construct generated dsRNA hairpins as a key elicitor for gene silencing (Christophe et al., 2003), the recombinate vector pTY-BrPDS was transformed into plants to confirming the visual success of VIGS. After three weeks of inoculation, photo-bleaching was observed in the leaves of pTY-BrPDS plant (Fig. 4A). Total RNA was extracted from the diseased ‘Bre’ leaves and reverse-transcribed into cDNA, which was used for the template of PCR with specific primers of virus coat protein (CP) gene. By sequencing, we verified that the CP mRNA was significantly abundant in the diseased ‘Bre’ plants (Fig. 5A); which implicated that TYMV virus had successfully infected Chinese cabbage.
Table 3: Sequences of the specific primers used in quantitative real-time PCR.
| gene name | Sense Primer | Anti-sense Primer |
| CP | TCCACCCTCACCACCTTC | GGGACAGACCTCGCTAACT |
| BrULT1 | ATGTGACCAAGACAAGTT | TTCTCCTCCAACGATAAC |
| BrULT2 | GTGTTCATTGAGGGAGAC | CCGTTGGAGTTATTCTTCA |
| BrActin | TTGCTATTCAGGCTGTTCT | CACCATCACCAGAGTCAA |
Twenty days post-inoculation, the heading type ‘Bre’ infected with pTY-BrULT2 exhibited an obviously irregulate growth state with an increased number leaves comparing with control plant (pTY-S), showing a distinctive phenotype with smaller and curled rosette leaves (Fig. 4A). The non-heading type ‘49 caixin’ infected with pTY-BrULT2 virus also showed abnormal phenotypes along with slender and irregularly curved stem, additionally the leaves number increased significantly with an increased branch outgrowths (Fig. 4A). To confirm the silencing efficiency at molecular level, the BrULT2 gene transcripts in the leaves of heading type ‘Bre’ was investigated by reverse transcription qPCR. The analysis result showed that the transcription level of BrULT2 gene dropped by 75% approximately, which indicated that the BrULT2 gene has been silenced successfully(Fig. 5B). Those novel phenotypes observed in the inoculated plant indicating that the ectopic BrULT2 expression alters the plant growth state leading to the modification of leaf shape of Chinese cabbage, additionally it also functions to regulate the stem and branch growth.
In present study, we also detected that the expression level of BrULT1 was up regulated 35% simultaneously when the BrULT2 gene of heading type ‘Bre’ was silenced (Fig. 5B). An interaction network predicated by using SMART software, as shown in Fig. 6, the BrULT2 protein was predicted to interact with BrULT1 and other proteins: GA, WUS, PAP7, SQN, RBL, TPR. So we hypothesis that the BrULT1 and BrULT2 gene interact genetically during the vegetative growth stage of Chinese cabbage, and the BrULT1 gene have similarly function with BrULT2 to regulate the rosette leaf development, with the pTY-BrULT2 mutants display abnormal phenotypes: curly leaves and ectopic branch outgrowth. The results were in line with many studies. For example, Monfared et al. (2013) reported that the ULT1 and ULT2 genes function redundantly to specify the apical-basal polarity axis of the gynoecium, indicating that they may function in differentiating tissues, and in a study by Monfared and Fletcher (2014) showed that the ULT2 gene play a minor but overlapping role with ULT1 in the regulation of shoot and floral stem cell accumulation.
Plant leaf shape is closely related to the photosynthetic efficiency and lodging resistance, exploring the leaf-shaped genes will contribute to the genetic improvement of excellent traits: disease resistance, stress resistance, high photosynthetic efficiency, leafy head formation. Previous report have demonstrated that class 1 KNOTTED1-LIKE HOMEOBOX(KNOX) genes are the common targets of ULT1 and ULT2 (Monfared et al. 2013), its ectopic expression may sculpt leaf morphogenesis with an auxin-directed mechanism controlling (Hay and Tsiantis, 2006). Nonetheless, future work will be required to exploring the hormone-related pathway of dramatic change in leaf shape for understanding the details of BrULT genes regulation.
Conclusion
In this study, we demonstrated that the optimized VIGS can be effectively performed to study specific gene function of Brassica rapa, and we firstly reveal that the BrULT2 gene act as an important epigenetic factor to regulating the leaf shape development of Brasssica rapa. Meanwhile, we hypothesis that the closely related BrULT1 and BrULT2 gene may play overlapping roles in the regulation of leaf curl status and other vegetative phenotypes. Our study provide some important information for exploring the specific functions of ULT family genes during vegetative stage of plants.
Acknowledgements
We thanked for providing Antoine Bouteilly (Centre National dela Recherche Scientifique) plasmid pTY-S. This work was supported by the grants from the Fundamental Research Funds for the Central University (KYZ201826), Jiangsu Agricultural Science and Technology Innovation Fund(CX(18)3068), and Jiangsu Agricultural Industry Technology System (JATS[2018]273).
References
- Alvarezvenegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). Atx-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Current Biology Cb, 13(8), 627-37.
- ÁlvarezBuylla ER, et al. (2015). The impact of polycomb group (pcg) and trithorax group (trxg) epigenetic factors in plant plasticity. New Phytologist, 208(3), 684-694.
- Becker, A., and Lange, M. (2010). VIGS-genomics goes functional. Trends in Plant Science, 15(1), 1-4.
- Bennypaul, H. S., Mutti, J. S., Rustgi, S., Kumar, N., Okubara, P. A., and Gill, K. S. (2012). Virus-induced gene silencing (vigs) of genes expressed in root, leaf, and meiotic tissues of wheat. Funct Integr Genomics, 12(1), 143-156.
- Bernadett Papp, and Jürg Müller. (2006). Histone trimethylation and the maintenance of transcriptional onand off states by trxg and pcg proteins. Genes Dev, 20(15), 2041-2054.
- Berr, A., et al. (2009). SET DOMAIN GROUP 25 encodes a histone methyltransferase and is involved in FLC activation and repression of flowering. Plant Physiol. 151, 1476–1485.
- Berr, A., et al. (2010). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell. 22, 3232–3248.
- Burch-Smith, T., Anderson, J., and Martin, G. K. S. (2010). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant Journal, 39(5), 734-746.
- Carles, C. C., Dan, C. I., Reville, K., Lertpiriyapong, K., and Fletcher, J. C. (2005). Ultrapetala1 encodes a sand domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development, 132(5), 897-911.
- Carles, C. C., and Fletcher, J. C. (2009). The sand domain protein ultrapetala1 acts as a trithorax group factor to regulate cell fate in plants. Genes & Development, 23(23), 2723-8.
- De, l. P. S. M., Acevesgarcía, P., Petrone, E., Steckenborn, S., Vegaleón, R., and Eshed, Y., Izhaki, A., Baum, S. F., Floyd, S. K., and Bowman, J. L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by kanadi and yabby activities. Development, 131(12), 2997-3006.
- Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. (2004). Validation of house-keeping genes for normalizing RNA expression in real-time PCR. BioTechniques 37:112–119.
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E. M., and Coupland, G. (1997). A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature, 386(6620), 44-51.
- Guo, L., Yu, Y., Law, J.A., and Zhang, X. (2010). SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl Acad. Sci. U S A. 107, 18557–18562.
- Hay, A., Barkoulas, M., and Tsiantis, M. (2006). Asymmetric leaves1 and auxin activities converge to repress Brevi pedicellus expression and promote leaf development in Arabidopsis. Development, 133(20), 3955-3961.
- Kant, R., Sharma, S., and Dasgupta, I. (2015). Virus-induced gene silencing (vigs) for functional genomics in rice using rice Tungro bacilliform virus (rtbv) as a vector. Methods in Molecular Biology, 1287, 201.
- Lacomme, C., Hrubikova, K., and Hein, I. (2003). Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant Journal, 34(4), 543–553.
- Manhães, A. M. E. D. A., Oliveira, M. V. V. D., and Shan, L. (2015). Establishment of an efficient virus-induced gene silencing (vigs) assay in Arabidopsis by agrobacterium-mediated rubbing infection. Methods in Molecular Biology, 1287(02), 235
- Maria, D. L. P. S. , Aceves-García, Pamela, Petrone, E. , Steckenborn, S. , Vega-León, Rosario, and álvarez-Buylla, Elena R., et al. (2015). The impact of polycomb group (pcg) and trithorax group (trxg) epigenetic factors in plant plasticity. New Phytologist, 208(3), 684-694.
- Martínezherrera, D., Romero, J., Martínezzapater, J. M., and Ponz, F. (1994). Suitability of Arabidopsis thaliana as a system for the study of plant-virus interactions. Fitopatología.
- Monfared, M. M., Carles, C. C., Rossignol, P., Pires, H. R., and Fletcher, J. C. (2013). The ult1 and ult2 trxg genes play overlapping roles in Arabidopsis development and gene regulation. Molecular Plant, 6(5), 1564-79.
- Monfared, M. M., and Fletcher, J. C. (2014). The ult trxg factors play a role in Arabidopsis fertilization. Plant Signaling & Behavior, 9(12), e977723.
- Mysore, K. S. (2011). New dimensions for vigs in plant functional genomics. Trends in Plant Science, 16(12), 656-65.
- Papp, B., and Müller, J. (2006). Histone trimethylation and the maintenance of transcriptional on and off states by trxg and pcg proteins. Genes & Development, 20(15), 2041-2054.
- Pires, H. R., Monfared, M. M., Shemyakina, E. A., and Fletcher, J. C. (2014). Ultrapetala trxg genes interact with kanadi transcription factor genes to regulate Arabidopsis gynoecium patterning. Plant Cell, 26(11), 4345-61.
- Pires, H. R., Shemyakina, E. A., and Fletcher, J. C. (2015). The ultrapetala1 trxg factor contributes to patterning the Arabidopsis adaxial-abaxial leaf polarity axis. Plant Signaling & Behavior, 10(7).
- Saleh, A., et al. (2008). The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell. 20, 568–579.
- Schmittgen, T. D., & Livak, K. J. (2008). Analyzing real-time pcr data by the comparative c(t) method. Nature Protocols, 3(6), 1101-1108.
- Tamada, Y., Yun, J.-Y., Woo, S.C., and Amasino, R. (2009). ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell. 21, 3257–3269.
- Tsiantis, M., & Langdale, J. A. (1999). Disruption of auxin transport is associated with aberrant leaf development in maize. Plant Physiology,121(4), 1163-1168.
- Vaghchhipawala, Z., Rojas, C. M., Senthil-Kumar, M., and Mysore, K. S. (2011). Agroinoculation and agroinfiltration: simple tools for complex gene function analyses. Methods in Molecular Biology, 678, 65.
- Velásquez, A. C., Chakravarthy, S., and Martin, G. B. (2009). Virus-induced gene silencing (vigs) in Nicotiana benthamiana and tomato. Journal of Visualized Experiments, 28(28), : 1292.
- Ye, J. , Chua, N. H. , Qu, J. , Geng, Y. F. , and Bu, Y. P. . (2014). Virus induced gene silencing (vigs) for functional analysis of genes in cotton.
- Yu, J., Yang, X. D., Wang, Q., Gao, L. W., Yang, Y., and Xiao, D., et al. (2018). Efficient virus-induced gene silencing in Brassica rapa, using a turnip yellow mosaic virus vector. Biologia Plantarum, 1-9.
- Zheng, B., and Chen, X. (2011). Dynamics of histone h3 lysine 27 trimethylation in plant development. Current Opinion in Plant Biology,14(2), 123-129.