Department of Biotechnology, K. L.E. Technological University, Hubballi, Karnataka, India
Corresponding author email: hombalimath@kletech.ac.in
Article Publishing History
Received: 09/12/2020
Accepted After Revision: 20/03/2021
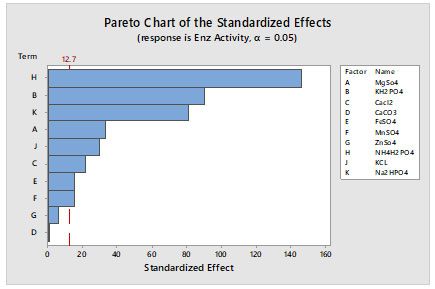
After performing OFAT analysis, screening of media component and process parameters was performed by PB design. Process and media optimisation of lipase are significant in view of economic production of enzymes in industries. Present study emphasises on optimising the process parameters for lipase production by bacteria through submerged fermentation using statistical tools. Bacterial strains were isolates from oil contaminated soil sources and screened for their lipase production based on the zone of clearance on Tributyrin Agar media. Eight isolates showing good ability to produce lipase were characterized for morphological features and motility. Further, one isolate was chosen and screened for media components (mineral salts) for lipase production by Plackett-Burman (PB) Design using Minitab 16 software. Totally eleven different minerals salts with 2 levels were considered for screening by PB design and 12 experimental design were taken for the study. The Pareto chart showed that except G (ZnSO4) and D (CaCO3) remaining all mineral salts showed significant effect on lipase production.
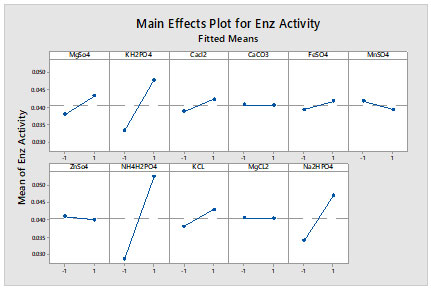
Main effect plots for enzyme production also found that for some of salts like CaCO3 and MgCl2 their change in concentration from low to high level does not any significance on enzyme production. Lipase activity ranged from 23U/ml to 72.3U/ml. NH4H2PO4 was found to be the most significant salt of the media component affecting the lipase production having P-value less than 0.05. A Full factorial design (FFD) was employed to determine the effects of media components (molasses, peptone and NH4H2PO4). The results of FFD show that 1.5ml for molasses; 0.2grams of NH4H2PO4 and1 gram of peptone were the optimized values. There exist significant interactions between molasses and peptone and NH4H2PO4 and peptone. The analysis of variance (ANOVA) was performed to check the adequacy of the proposed models. The results of RSM based mathematical modelling, indicates the ability of this technique to predict proposed performance at 95% confidence interval. The coefficient of determination R-sq was 97.11%, which confirms that the model was statistically significant and good fit.
Lipase; Mineral Salts; Pb-Design; Full Factorial Design; Submerged Fermentation.
Hombalimath V. S, Kamaraddi J. A, Sharanappa A, Desai S. V, Shet A. R, Patil L. R. Statistical Optimization of Lipase Production from Bacillus species by Submerged Fermentation. Biosc.Biotech.Res.Comm. 2021;14(1).
Hombalimath V. S, Kamaraddi J. A, Sharanappa A, Desai S. V, Shet A. R, Patil L. R. Statistical Optimization of Lipase Production from Bacillus species by Submerged Fermentation. Biosc.Biotech.Res.Comm. 2021;14(1). Available from: <a href=”https://bit.ly/2LqjNdQ”>https://bit.ly/2LqjNdQ</a>
Copyright © Hombalimath et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Lipases (triacylglycerol acylhydrolases, EC 3.1.1.3) are enzymes that are unique in catalyzing the hydrolysis of fats into fatty acids and glycerol at the water-lipid interface, and have the ability of reversing the reaction in non-aqueous media. The recent applications of lipases are additives in food (flavour modification), fine chemicals (synthesis of esters, detergent hydrolysis of fats), wastewater treatment (decomposition and removal of oily substances), cosmetics (removal of lipids), pharmaceuticals (digestion of oil and fats in foods), leather processing (removal of lipids from animal skins) and biomedical assays (blood triglycerides). (Manouchehr et al., 2018). Additionally, lipases have an important application in the field of bio energy, especially in biodiesel production, which is an expanding sector, as a result of the worldwide rising demand for the use of renewable energy (Daiha et al., 2015; Manouchehr et al., 2018).
Lipases are carboxyl esterase’s that catalyze the hydrolysis of acylglycerols containing fatty acid chains greater than 10 carbon atoms in length. There is a great to militate these effluents which has necessitated the development of new technology for bioremediation involves the intervention of microorganism to degrade the pollutants through these metabolites like enzymes (Daiha et al., 2015). Various microorganisms producing lipase are used in bioremediation of oil contaminated soil. Hence lipases are regarded as potential agent for commercial applications. They are being explored for both academically and industrially for their applications. Lipids are essential components of living systems and are important sources of energy.
They are part of cell membranes and signalling events. Lipids are needed to catalyze these reactions (Gilham and Lehner, 2005; Mateus et al., 2009; Salihu et al., 2011; Karina et al., 2016).The enzyme from microbial sources is currently receiving more attention because of their potential diverse applications in industries such as detergent, oleo chemical, organic. Considering the fact that a substantial part of industrial enzymes production cost is contributed by the cost of the fermentation medium, the present investigation was aimed at evaluating the effects of medium components on lipase production by formulating a suitable medium containing waste groundnut oil as a low-cost renewable substrate and molasses as an economic carbon source (Pin et al., 2015; Nelison et al., 2016). Plackett-Burman (PB) design is important statistical design used for the screening and selection of medium components in shake-flask cultures.
PB design offers a good and fast screening procedure and it mathematically computes the significance of a large number of factors in one experiment.It has been applied to try and reduce the number of fermentations runs to an absolute minimum. It saves much of the time and gives useful information on each component using a minimum number of experiments compared to conventional method of screening (Parimala et al., 2006; Salihu et al., 2011; Selvam et al., 2016).It is used only to evaluate and select the significant factors, which influence the production of lipase during fermentation. It does not describe the interactions among the factors affecting the process. Full Factorial Design (FFD) provides the most complete information, it often requires so many runs that they become impractical to carry out.
Thus, optimum performance has been determined using mathematical tools such as multiple regression of a partial or full factorial to obtain a model of the production system, usually involving fitting of data to a polynomial equation, using stepwise multiple regression.Response surface methodology has also been used to investigate the optimal regions of production of useful product. Detailed analyses of the optimized region using simple designs have also been applied for optimization processes. However, several interactions of the experimental design and optimization of models are required for effective application to product formation in fermentation systems (Jaeger et al., 1997; Savitha et al., 2007; Sirisha et al., 2010; Anwar et al., 2011; Pin et al., 2015).
Gap Analysis:Lipolytic enzymes are categorized into these groups- esterases, phospholipases and lipases are known to catalyze the hydrolysis of triglycerides into and free fatty acids and glycerol. Lipases usually act carboxylic bonds of triglycerides and give simple fatty acids and glycerol as by product. The key uniqueness of lipase is substrate specific; they are known to be stable at higher of pH and temperature. Lipases are extracellular in nature and act on substrate. Lipases having variety of applications are commonly regarded as third largest enzyme being produced only after protease and amylase. The present study caters the various needs of lipase enzyme as biocatalyst for different application.
MATERIAL AND METHODS
For the soil sample collection, the oil contaminated soil samples where soil was collected by digging ground up to 15-20 cm nearby Hubballi, with the help of clean spatula in sterile polythene bag. Immediately after collecting the soil it was stored in the refrigerator at 4 °C till processing (Pooja et al., 2015).For the isolation and screening of lipase producing bacteria, 5 grams of the soil sample was added to nutrient broth (100ml.) and incubated for 24 hours. Soil sample was serially diluted (0.85% NaCl) and plated on tributyrin agar base containing 0.5% (w/v) peptone, 0.3% (w/v) beef-extract, 1% (v/v) Tributyrin and 2% agar, pH 7.0 by spread plate method. Plates were incubated at 37 °C for two days.
Pure cultures of the isolates were maintained on nutrient agar slants supplemented with tributyrin. The isolated colonies were picked and maintained on nutrient agar slants as pure cultures. These were screened for lipolytic activity by Tributyrin Qualitative Plate Assay based on the zone of clearance (by incubating at 37 °C for 2 days). Isolates showing maximum zone of clearance were taken for further studies. Morphological characterization of these isolates was performed by gram staining technique and Motility test (Hanging drop technique) (Rifaat et al., 2010; Acikel et al., 2011; Veeranna et al., 2012).For the microbial culture and inoculum preparation, one isolate having maximum lipase activity was chosen for further studies.
Loop full of the culture was inoculated into the inoculum media(100ml) containing molasses(1.5ml), waste groundnut oil (2.2ml), peptone(1g), CaCO3(0.1g), (NH4)2SO4(0.1g), MgSO4.7H2O (0.1g), Inoculum flask was kept for incubation for 2 days at 37°C and 150rpm (Imandi et al., 2007). For the lipase production media, 1ml of inoculum in 100ml production medium containing Soya peptone (1gram), waste groundnut oil (2.2ml), KH2PO4 (0.5gram), CaCO3 (0.1gram), (NH4)2SO4 (0.1gram), MgSO4.7H2O (0.1gram), Molasses (1.5gram) and submerged fermentation was carried out at 37oC 150 rpm for 72 h (Imandi et al., 2007). For the fermentation medium and lipase production for screening of mineral salts, the fermentation medium was prepared by adding molasses (1.5ml), waste groundnut oil (2.2ml), peptone (1gram) and 11 combinations of mineral salts and inoculum was added to fermentation medium.
For the full factorial design, in order to evaluate the factors that influence the lipase production using cheaper carbon source, a three level and three factor full factorial experiments was designed. Carbon source, nitrogen source and NH4H2PO4 (which showed most significance by Pareto chart P (<0.005)) were chosen as the independent variables and lipase activity (U) was taken as the response of the design. The level and the range of the independent variables were examined in three levels. Thus, ranges of (0.5, 1.5 and 2.5%v/v) of molasses as carbon source, (0.5, 1 and 1.5%v/v) of peptone as nitrogen source and (0.2, 1, 1.8%v/v) of NH4H2PO4 were considered. Practical considerations were used for selecting the three factors and the range in which they were varied (Prashant et al., 2006; Anwer et al., 2011; Ananthi et al., 2013).
RESULTS AND DISCUSSION
Isolation and Screening of bacterial strains: Many bacterial colonies were isolated from oil contaminated soil and screened for lipase production by tributyrin zone of clearance test. Totally eight isolates showing the maximum zone of clearance was chosen for morphological studies (Ajit et al., 2007; Bhavani et al., 2012).
Figure 1: Tributyrin Agar media
Selection of significant mineral salts by PB Design: PB design was used to evaluate maximum lipase production as function of mineral salts. A total of eleven medium components (minerals) were studied with regard to their effects on lipase production by PB design using the Minitab16 software. The combinations of medium components (minerals) are shown in [Table 1]. Based on the design matrix selected for the screening of significant variables, the highest lipase activity realized was 72.3 U/ml at run 1 and the lowest amount was observed in run 2 (23 U/ml) (Sirisha et al., 2010; Bhavikatti et al., 2020).
Table 1. PB design matrix for screening mineral sources
| S. No | MgSO4.7H2O | KH2PO4 | CaCl2 | CaCO3 | FeSO4 | MnSO4 | ZnSo4 | NH4H2P04 | KCl | MgCl2 | NaH2PO4 | Enzyme Activity (U) | Lipase con(µg/ml) |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 72.3 | 0.2661 |
| 2 | -1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | 23 | 0.0946 |
| 3 | -1 | -1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | 24.3 | 0.3534 |
| 4 | 1 | -1 | -1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 34 | 0.1311 |
| 5 | -1 | 1 | -1 | -1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | 53.3 | 0.1257 |
| 6 | -1 | -1 | 1 | -1 | -1 | 1 | -1 | 1 | 1 | 1 | -1 | 38 | 0.3364 |
| 7 | -1 | -1 | -1 | 1 | -1 | -1 | 1 | -1 | 1 | 1 | 1 | 25.3 | 0.0369 |
| 8 | 1 | -1 | -1 | -1 | 1 | -1 | -1 | 1 | -1 | 1 | 1 | 53 | 0.0851 |
| 9 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | -1 | 1 | -1 | 1 | 44 | 0.1475 |
| 10 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | -1 | 1 | -1 | 31 | 0.2972 |
| 11 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | -1 | 1 | 63.3 | 0.2633 |
| 12 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | -1 | 1 | -1 | -1 | 24.67 | 0.2828 |
Figure 2: Pareto graph showing variables effect on maximum lipase production, based on PB design observations
Figure 3: Main effects plot for enzyme activity
Response Surface Methodology (RSM)/Full Factorial Design (FFD) for media components: Full factorial Design is used to determine the optimum response of lipase production. FFD for three independent variables was used to obtain the combination of values that optimizes the response. The experiments were designed using the Minitab16 software. Based on the PB design results further studies was done by performing RSM to optimise the lipase production using significant media components. The significant independent variables of the medium components are molasses (carbon source), peptone (nitrogen source) and NH4H2PO4. FFD experiment matrix along with response is shown in following table.
The ANOVA table obtained from analyzing the response we got the value of R-Sq (pred) and R-Sq (adj) difference is less. Hence, the designed model is adequate and consistent. Analysis of variance showed a high coefficient of determination of (R2) value of 97.11%, indicating a satisfactory fit of the model with the experimental data. All the three parameters were statistically significant at p<0.05 and all the parameters influence the production of lipase as shown in the Pareto chart (Kumar et al., 2006; Rao et al., 2010; Tembhurkar et al., 2012; Muhhamad et al., 2012; Bhavikatti et al., 2020).
Table 2. FFD matrix showing effect of media components on lipase activity
| Molasses | NH4H2PO4 | Peptone | Response-EA(U/ml) | |
| -1 | -1 | -1 | 10.21 | |
| -1 | 0 | -1 | 5.11 | |
| -1 | 1 | -1 | 3.92 | |
| 0 | -1 | -1 | 13.89 | |
| 0 | 0 | -1 | 13.59 | |
| 0 | 1 | -1 | 9.85 | |
| 1 | -1 | -1 | 12.01 | |
| 1 | 0 | -1 | 9.13 | |
| 1 | 1 | -1 | 6.84 | |
| -1 | -1 | 0 | 9.28 | |
| -1 | 0 | 0 | 6.69 | |
| -1 | 1 | 0 | 5.41 | |
| 0 | -1 | 0 | 18.64 | |
| 0 | 0 | 0 | 15.90 | |
| 0 | 1 | 0 | 12.86 | |
| 1 | -1 | 0 | 9.87 | |
| 1 | 0 | 0 | 9.36 | |
| 1 | 1 | 0 | 7.67 | |
| -1 | -1 | 1 | 8.54 | |
| -1 | 0 | 1 | 5.14 | |
| -1 | 1 | 1 | 10.98 | |
| 0 | -1 | 1 | 13.67 | |
| 0 | 0 | 1 | 11.24 | |
| 0 | 1 | 1 | 11.12 | |
| 1 | -1 | 1 | 6.48 | |
| 1 | 0 | 1 | 4.37 | |
| 1 | 1 | 1 | 8.58 | |
Analysis of Variance:
| Source | DF | Seq SS | Contribution | Adj SS | Adj MS | F-Value | P-Value |
| Model | 18 | 335.554 | 97.11% | 335.554 | 18.642 | 14.94 | 0.000 |
| Linear | 6 | 253.536 | 73.38% | 253.536 | 42.256 | 33.87 | 0.000 |
| Molasses | 2 | 196.996 | 57.01% | 196.996 | 98.498 | 78.95 | 0.000 |
| NH4H2PO4 | 2 | 42.259 | 12.23% | 42.259 | 21.130 | 16.94 | 0.001 |
| Peptone | 2 | 14.281 | 4.13% | 14.281 | 7.141 | 5.72 | 0.029 |
| 2-Way Interactions | 12 | 82.017 | 23.74% | 82.017 | 6.835 | 5.48 | 0.011 |
| Molasses*NH4H2PO4 | 4 | 11.364 | 3.29% | 11.364 | 2.841 | 2.28 | 0.150 |
| Molasses*Peptone | 4 | 30.972 | 8.96% | 30.972 | 7.743 | 6.21 | 0.014 |
| NH4H2PO4*Peptone | 4 | 39.681 | 11.48% | 39.681 | 9.920 | 7.95 | 0.007 |
| Error | 8 | 9.981 | 2.89% | 9.981 | 1.248 | ||
| Total | 26 | 345.535 | 100.00% |
Regression Equation
Response-EA (U/ml) = 9.643 – 2.391 Molasses_-1 + 3.776 Molasses_0 – 1.385 Molasses_1
+ 1.756 NH4H2PO4_-1 – 0.694 NH4H2PO4_0 – 1.062 NH4H2PO4_1
– 0.248 Peptone_-1 + 0.989 Peptone_0 – 0.740 Peptone_1
CONCLUSION
Eleven mineral salts were screened by 12 experimental runs. The experimental plan and corresponding lipase activity were reported. The Pareto chart were used to show the effect of all the mineral salts on lipase production. The Pareto chart showed that except G (ZnSO4) and D (CaCO3) remaining all mineral salts showed significant effect on lipase production. Out of these NH4H2PO4 with P- value (0.004), KH2PO4 with P value (0.007) and NaH2PO4 with P– value (0.008) showed major effect on lipase production. In addition, predicted R2 of PB design was 99.63%, which is a good model fit. Three factors were considered at 3 levels for FFD, total 27 experiments were performed to evaluate the enzyme activity. FFD results showed that all independent variables are significant having the P value less than 0.05 (except molasses and NH4H2PO4 interaction). From FFD we got 1.5ml for molasses, 0.2gram for NH4H2PO4 and 1gram peptone as optimum value. There exist significant interactions between molasses and peptone and NH4H2PO4 and peptone. R-sq of FFD design was 97.11% which confirm that our model is good fit.
REFERENCES
Acikel, U, Ersan, M and Sag-Acikel, Y. (2011). The effects of the composition of growth medium and fermentation conditions on the production of lipase by rhyzopusdelemar. Turkey Journal of Biology. 35:35-44.
Ahmed, E. H., Raghavendra, T, and Madamwar, D. (2010). An alkaline lipase from organic solvent tolerant acinetobacter species. EH28: application for ethylcaprylate synthesis. Journal of Bio resources and Technology.101: 3628 – 3634.
Anwar T., Mathur A., Mathur G., and Chauhan R.S., (2011) Statical optimization for co- production and partial characterization of thermo-halotolerans lipase and α-amylase from halotolerans Bacillus subtitles JPBW-9. World Journal of Science Technology. 1: 54-63.
Anwar T., Mathur A., Mathur G., and Chauhan R.S., (2011) Statical optimization for co- production and partial characterization of thermo-halotolerant lipase and α-amylase from halo tolerant Bacillus subtilis JPBW-9. World Journal of Science Technology. 1: 54-63.
Babu, I.S., Chisti, Y., and Banerjee, U.C., (2007), Lipase production by yarrowia lipolytica in solid state fermentation using mixed substrate, Research Journal of Microbiology, 2 (4): 469- 474
Bhavani M., Chowdary, G.V., David M., and Archana G., (2012). Screening, Isolation and Biochemical Characterization of Novel Lipase Producing Bacteria from Soil Samples. International journal of biological engineering. 2: 8-22.
Bhavikatti, J., Bodducharl, S., Kamagond, R., Desai, S., and Shet, (2020 A). Statistical Optimisation of Protease Production Using a Freshwater Bacterium chryseobacterium cucumeris SARJS-2 for Multiple Industrial Applications. Journal of 3 Biotech, 10, (6).
Daiha, K.D.G., Angeli, R., de Oliveira, S.D. and Almeida, R.V., (2015). Are lipases still important biocatalysts? A study of scientific publications and patents for technological forecasting. PloS one, 10(6), p.e0131624.
de Godoy Daiha, K., Brêda, G.C., Larentis, A.L., Freire, D.M.G., and Almeida, R.V., (2016). Enzyme technology in Brazil: trade balance and research community. Brazilian Journal of Science and Technology, 3(1), pp.1-13.
Gilham, D. and Lehner, R. (2005). Techniques to Measure Lipase and Esterase Activity in vitro. methods, Journal of advances in Enzyme Research. 36:139-147.
Godoy, K. Daiha, R. Angeli, Dias S. L., Rodrigo, and Almedia, (2015). Are Lipases Still Important Biocatalysts? A Study of Scientific Publications and Patents for Technological Forecasting, Journal of pone.0131624 PLOS ONE10 (6): e 0131624. doi: 10.1371.
Hombalimath, V.S., Desai, V.S., and Sharanappa A., (2020). Characterization of lipase immobilized on Chitosan magnetic micro-particles for economic biodiesel production. International Journal of Scientific and Technological Research, 9(3): 5111-5116.
Hombalimath, V.S., Udapudi, B.B., Patil, L.R., Shet, A.N., Yaraguppi, D.A. and Tennalli, G., (2012). Isolation and characterization of lipolytic microorganisms from oil contaminated soil. Int J Adv Sci Eng Technol, 2(3), pp.293-297.
Jaeger, K.E., (1997), Bacterial lipases for biotechnological applications, Journal of Molecular Catalysis B: Enzymatic, vol-3, pg 3-12.
Javed, M.M., (2007). A 23 Level full factorial design for optimization of cultural conditions for lipase production by consortium of Aspergillus Niger and Trichoderma Virdde. Research Journal of Microbiology, 2 (8): 639-644.
Javed, M.M., (2012). Optimization of cultural conditions for lipase production by submerged culture of Rhizopus Oligosporus TUV-31, Pakistan Journal of Botany. 35(4): 519-525
Kandasamy, S., Muthusamy, G., Balakrishnan, S., Duraisamy, S., Thangasamy, S., Seralathan, K.K., and Chinnappan, S., (2016). Optimization of protease production from surface-modified coffee pulp waste and corncobs using Bacillus sp. by SSF. 3 Biotech, 6(2), pp.1-11.
Kumar A., Parihar, S.S., and Batra, N., (2007). Enrichment, isolation and optimization of lipase-producing staphylococcus species. from oil mill waste (Oil cake). Journal of Experimental Sciences. 3(8): 26-30.
Kumar, M.P.P., and Valsa, A. K., (2006), Optimization of culture media and cultural conditions for the production of extracellular lipase by bacillus coagulans, Indian journal of biotechnology, vol-6, pg 114-117.
Mahale, P.K., Desai, S.V., Hombalimath, V.S. and Achappa, S., (2014). Isolation, screening and characterization of lipase producing strain from oil contaminated soil of Hubballi, Karnataka. International Journal of Basic and Applied Biology (IJBAB), pp:198.
Neilson, P.M., (2016). Production of Biodiesel Using Liquid Lipase Formulations and Chemical Engineering Society 93:905–910, Springer Open source.
Rao, P., and Divakar, S. (2001), Lipase catalyzed etherification of terpineol with various organic acids: application of the Plackett–Burman design, Process Biochemistry, vol :(36), pp: 1125-1128.
Salleh, A.B., Musani, R., Basri, M., Ampon, K., Yunus, W.M.Z., and Razak, C.N.A., (1993). Extra-and intra-cellular lipases from a thermophilic Rhizopus oryzae and factors affecting their production. Canadian journal of microbiology, 39(10), pp.978-981.
Tembhurkar V. R., Kulkarni M. B., and Peshwe S. A., (2012). Optimization of Lipase Production by Pseudomonas sp. in submerged batch process in shake flask culture. Journal of science research reporter. 2: 46-50.