1Rayalaseema University, Kurnool, India
2Dr. Bidlan’s Research Institute, Hyderabad, India
2Department of Biotechnology, Delhi Technological University, Shahbad Daulatpur, Delhi-110042
Article Publishing History
Received: 02/01/2018
Accepted After Revision: 19/03/2018
Organochlorine pesticides (OCPs) primarily 1, 1, 1-trichloro-2,2-bis(4-chlorophenyl) ethane (p,p’-DDT) and ã-hexachlorocyclohexane (lindane) pose adverse health effects to the environment and community. Lindane and DDT (dichlorodiphenyltrichloroethane) have been broadly used for agricultural purposes. DDT is still the “sought after” for public health care programs to control vector-borne diseases like malaria in developing nations. Even though both these compounds are prohibited in many parts of the world, their residues are still found in various environmental, food, human and animal samples. In this study, the microbial population isolated from aquatic systems, rivers from Yamuna (North India) and Godavari (South India) was enriched until a Lindane and DDT tolerant population was established. Screening of the population for understanding bioremediation potential was done using 5ppm of DDT and Lindane. The populated microbial cells formed the consortium that was used subjected to metagenomic analysis to identify the organisms till species level. The 16S amplicon sequences were analysed using Kaiju online tool for establishing the identity of individual bacteria present in the mixed population. This consortium was shown to consist of 138 species with 11 uncultured bacteria with Taylorella asinigenitalis holding the maximum numbers in its population. The microbial consortium was found simultaneously degrading DDT and Lindane in shake flasks. The enriched consortium could degrade 95% of 5 ppm lindane and 57.8% of 5 ppm DDT in the supplied mixture by the end of 72 h. This is a promising observation and the consortium shows high efficiency in degrading the OCPs. The consortium can become a vital tool for biodegradation of organochlorine pesticides to eradicate the pesticide residues in water and soil ecosystems.
Microbial Consortium, Biodegradation, Kaiju, Ddt, Lindane
Saghee M. R, Bidlan R. Simultaneous Degradation of Organochlorine Pesticides by Microbial Consortium. Biosc.Biotech.Res.Comm. 2018;11(1).
Saghee M. R, Bidlan R. Simultaneous Degradation of Organochlorine Pesticides by Microbial Consortium. Biosc.Biotech.Res.Comm. 2018;11(1). Available from: https://bit.ly/2MSxK2f
Introduction
1,1,1-trichlorão-2,2-bis(4-chlorophenyl) ethane (p,p’-DDT) and -hexachlorocyclohexane (lindane) are the two most extensively applied pesticides in the later part of the 20th century by many nations globally (Aktar et al. 2009, Donald, 2001). Even though both these compounds are prohibited in many parts of the world, their residues are still found in environment, food, human and animal samples. Lindane has been enlisted under carcinogenic compounds while DDT is a suspected oncogenic agent. The water bodies, agricultural soil and the glaciers in the polar regions of the earth show the presence of these synthetic pesticides. It has become a priority issue for the scientific fraternity to clear off these residues from the environment ultimately leading to pesticide-free food, water, fruits, beverages, vegetables, etc. and providing the future generations a conducive condition to thrive, (Pandey et al. 2011, Villa et al. 2003, Wilson et al. 2013, Loomis et al. 2015, Muinck, et al. 2017 and Jayaraj et al. 2017).
Lot of research has been carried out to answer this problem, but not much clearance of the pesticide residues is documented in the field applications (María et al. 2010). Even though there are many processes that have been protected under patents (Bidlan et.al. 2009) often no efforts are made to create awareness among the government agencies, farmers and others to exploit them. Most of the research in bioremediation revolves around a few microbes and a single compound at the laboratory level. Only the effluent treatment plants use the natural microbial flora in their systems. Still no organochlorine pesticide residues are eliminated from the nature leading to their persistence and causing major health issues among the population. In this paper, we describe a diverse population of microbes in a consortium isolated from Indian rivers Yamuna and Godavari and characterised using 16S metagenomics through illumina platform (Muinck, et al. 2017) that demonstrates the potential to degrade the organochlorine insecticides DDT and lindane provided as a mixture in shake flasks
Material and Methods
Chemicals
99.4% pure p,p’-DDT was donated for research by Hindustan Insecticides Ltd., India while 97% pure lindane was purchased from Sigma Aldrich, USA. HPLC grade acetone and ethyl acetate was procured from Merck. Other chemicals used were of analytical grades and purchased from standard manufacturers.
Microbial culture and enrichment: Water samples were collected from highly contaminated rivers Yamuna and Godavari. These samples were subjected to increasing concentrations of two organochlorine pesticides (OCP) 1,1,1-trichloro-2,2-bis-(4-chloro) ethane (DDT) and ã-hexachlorocyclohexane (lindane) over a period of 6 months. Periodically it was tested for the viability of the population through streaking on to nutrient agar (Bidlan, 2003).
Screening for the OCP degradation potential: The microbial population developed through enrichment was screened for its potential to degrade DDT and lindane mixture simultaneously. 25 mL of sterile water through reverse osmosis and autoclaving was spiked with a mixture of 5 ppm DDT and Lindane. The enriched microbial population was inoculated to this and incubated at ambient condition in a rotary shaker set at 150 rpm. Samples were drawn periodically and analysed for residual OCPs (Bidlan and Manonmani 2004).
Extraction of OCPs: Samples drawn at every 24 h were acidified and extracted with equal volume of dichloromethane twice and organic layers pooled after passing through anhydrous sodium sulphate and fluorisil. The residual OCPs were transferred into microfuge tubes after completely drying the dichloromethane and dissolving in a small volume of acetone.
Thin Layer Chromatography: TLC was performed on 0.25 mm thick silica gel G layers. Residual OCPs were dissolved in a known volume of acetone. Equal volumes of this solution carrying 10 micrograms of each pesticide at 0 h were loaded and developed in a saturated environment of cyclohexane. The air-dried plates were sprayed with 2% o-tolidine solution in acetone under bright sunlight to detect the individual OCP spots. The intensity and area under each spot was considered to quantify the residual insecticides by using the standard curve plotted with vs log (Concentration).
Gas Chromatography-Mass Spectrometry Fingerprinting: Qualitative and quantitative analyses were confirmed with GCMS/MS fingerprinting using Agilent 7000D equipment with a triple quad (Gawad, 2016). The column HP-5ms (Agilent 19091S EPC) was programmed with pressure 30.797 psi, the flow of 3.1793 mL.min-1, average Velocity of 54.506 cm.s-1 and temperature 70°C to 280°C, the sample was electron ionized (EI) with a source temperature of 300°C. The quantification was done using a standard curve prepared for different amounts of the two OCPs (DDT and Lindane) under the same conditions (Muir, 2006).
Genomic DNA extraction: DNA was isolated using Xcelgen Bacterial gDNA kit with few modifications. Quality of gDNA was checked on 0.8 % agarose gel and quantification was done on Nanodrop 8000.
Microbial Identification: The microbial population was tested for the presence of various bacteria using 16S rRNA gene analyses through metagenomic approach. Illumina platform (Blomquist TM et al. 2013) was used to carry out next-generation sequencing of ~460 bp amplicons obtained using Prokaryotic V3 Forward primer 5’ CCTACGGGNBGCASCAG3’ and Prokaryotic V4 Reverse primer 5’ GACTACNVGGGTATCTAATCC3’. The obtained 1,94,544 sequences were subjected to online tool Kaiju (http://kaiju.binf.ku.dk/server) for identification purpose.
Results and Discussion
The long-term enrichment of the water sample with the mixture of DDT and Lindane resulted in a microbial population that contained many types of microorganisms, including bacteria and fungi. Some of the strains were isolated by repeated streaking. The degradation of mixture of DDT and Lindane, both at 5 ppm concentrations was reflected in the TLC plate (Figure. 1) and confirmed by GCMS/MS fingerprinting. There was a marked reduction in the residual concentration of Lindane in three days of incubation. The amount of DDT also reduced from 0 h to 72 h. The visual/apparent concentrations in the spots for both the insecticides is a clear indication that the microbial population was effectively working against the OCPs supplied and catabolising these compound for its growth and energy. Since the only source of carbon in the medium was the mixture of DDT and Lindane, the other source of carbon can be the atmospheric carbon in the form of carbon dioxide and few cell masses that must have died. There was an increase in the biomass with incubation, indicating the efficiency of the consortium to resist the OCP and grow in sole supply of these compounds as carbon source. However, the TLC clearly indicated the degradation of DDT and Lindane in presence of the consortium.
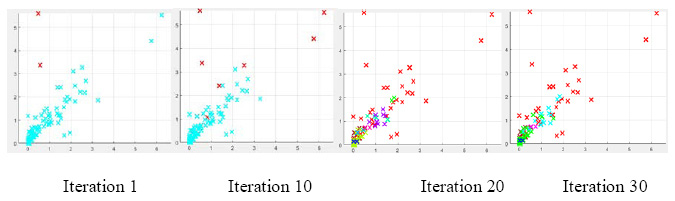
Gas Liquid Chromatograms show that the retention time of Lindane was 13.3 min while that of p,p’-DDT was 26.9 min under the conditions of analyses. Fig. 3 gives the chromatograms of the screening experiment with 5 ppm of each DDT and Lindane in the broth. There is a gradual decrease in the peak heights and area under them for both DDT and Lindane from 0 h to72 h. The degradation pattern of the two pesticides is given in Fig. 4. The enriched consortium could degrade 95% of 5 ppm Lindane and 57.8% of 5 ppm DDT in the supplied mixture by the end of 72 h. This is a promising observation and the consortium shows high efficiency in degrading the OCPs. Other research groups have also shown the degradation of DDT and Lindane by various individual microorganisms or a defined consortium having only a few strains (Bidlan, 2002). Simultaneous degradation of organochlorine and organophosphate pesticides was demonstrated by researchers (Abraham et al. 2014).
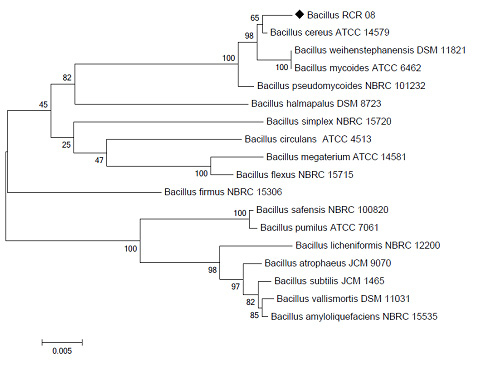 |
Figure 2: Gas Chromatograms of DDT and Lindane Residual Concentrations at different incubation periods at 0 Hr, 24 Hr, 48hr and 72 hr |
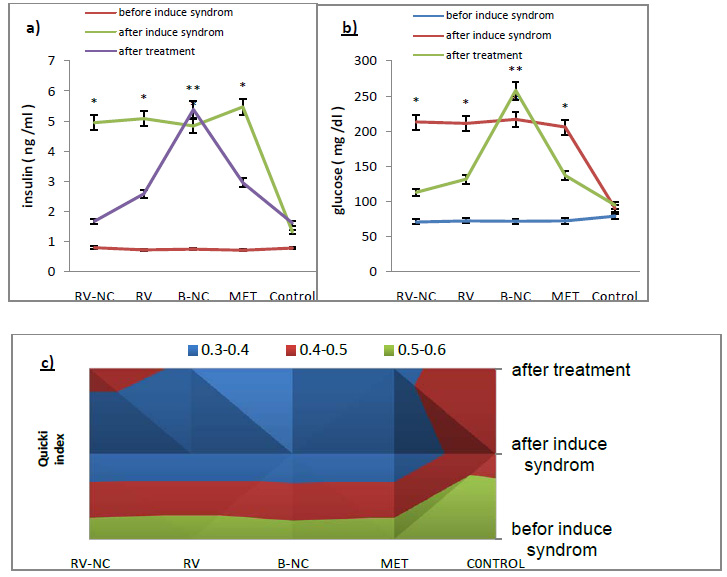 |
Figure 3: Degradation of DDT and Lindane by Consortium |
 |
Figure 4: Pie chart of species distribution of bacteria characterized using 16S metagenomics and Kaiju tool with relative abundance in the consortium |
This study is novel as it attempts to demonstrate the degradation of the mixture of DDT and Lindane (mixed organochlorines) by the bacterial consortium. This hold a great promise in future bioremediation applications.
Identification and relative abundance of various bacteria in the consortium
The consortium was subjected to metagenomic analysis through 16S rDNA gene sequencing using the set of prokaryotic primers. The amplified sequences of
approximately 146 bp were analysed after removing the chimeric reads. A total of 194544 reads were analysed by the webserver tool “Kaiju”. The program could identify 138 different species, few of which were uncultured bacteria. We have represented the top 63 species distribution in Fig. 5.
Taylorella asinigenitalis with 52.51 % relative abundance was the highest number of reads (128267). Second highest reads with a relative abundance of 8.13% was from Enterococcus faecalis. One uncultured bacterium was occupying the third position among the consortium population (Handelsman, 2004). A total of eleven uncultured bacteria/archaea were detected in the consortium out of which seven are present in the top 63. This report shows more than 100 various species of bacteria / archaea that might play a significant role in the bioremediation of OCPs. With more studies and analyses, the consortium can be further trained to degrade higher concentrations of these OCPs and other environmental threats caused by human activities. Complete DNA sequences obtained have been deposited at NCBI under the bioproject ID PRJNA420925.
Conclusion
Many reports have appeared in the last 6-7 decades regarding the use of microbes in cleaning of environmental contaminants. Ours is also an attempt with a view that the single axenic culture or just a few membered consortia might not suffice the need of the hour. Hence, a consortium developed from the highly contaminated river sources in India was demonstrated to have a great potential in remediating the environment off OCPs. This consortium was shown to consist of 138 species with 11 uncultured bacteria and Taylorella asinigenitalis holding the maximum numbers in its population. This consortium not only comprises of bacteria but also have various fungi that have not been considered in the present study. The complete analyses of this consortium and further studies to confirm its efficacy in treating higher amounts of OCPs and other compounds need to be taken up. This is just the beginning of a new era in combating the environmental and health issues, new avenues are yet to be explored. The consortium can become a promising solution for biodegradation of organochlorine pesticides to eradicate the pesticide residues in water and soil ecosystems.
Acknowledgements
The authors would like to acknowledge Rayalaseema University and UGC for support and encouragement for the research studies.
References
- Akhtar WM , Dwaipayan Sengupta, and Ashim Chowdhury (2009), Impact of pesticides use in agriculture: their benefits and hazards, Interdisciplinary Toxicology, 2(1), ,pp.1–12.
- Bidlan R, Manonmani H. K., Kunhi A.A.M. (2007). A process for degradation of dichlorodiphenyltrichloroethane (DDT) using an improved strain, PCT International Classification No. C02C 5/10
- Bidlan R. (2003), Studies on DDT degradation by Bacterial strains. In: Isolation, purification and identification of microbes capable of DDT- degradation. Ph.D. thesis, University of Mysore, India. pp 90-142.
- Bidlan R. and Manonmani H.K.(2004), Aerobic degradation of dichlorodiphenyltrichloroethane (DDT) by Serratiamarcescens DT-1P. Process Biochemistry,38, pp.49-56.
- Bidlan R., Manonmani H.K. (2007) A process for the enhanced degradation of Dichloro-diphenyltrichloroethane (DDT). Indian Patent. Application No. 520/DEL/2003A International Classification No. A01N 3/00
- Bidlan R., Manonmani H.K. (2007) A process for the preparation of biocatalysts useful for the degradation of dichlorodiphenyldichloroethylene (DDE). Indian Patent Application No. (522/DEL/2003A). International Classification No. B01J 37/36
- Bidlan R., Manonmani H.K. (2009), A process for the preparation of biocatalysts for the remediation of dichlorodiphenyldichloroethane (DDD/TDE) containing industrial effluents. Patent. 734/DEL/2005A, International Classification No. A01J
- Blomquist TM, Crawford EL, Lovett JL, Yeo J, Stanoszek LM. (2013), Targeted RNA Sequencing with Competitive Multiplex-PCR Amplicon Libraries. PLOS ONE 8(12): 10.1371
- David Wilson, Nageswara Rao, Narasimha Reddy, (2013), Concentration of Organochlorine pesticide residues in sediments from the Godavari River of East Godavari District of Andhra Pradesh, Journal of Chemical, Biological and Physical Sciences, May 2013- July; Vol. 3, No. 3; 2279-2292.
- Donald J.Ecobichon, (2001), Pesticide use in developing countries, Toxicology, Volume 160, Issues 1–3, 7 March, Pages 27-33
- Eric J. de Muinck, Pål Trosvik, Gregor D. Gilfillan, Johannes R. Hov and Arvind Y. M. Sundaram, (2017), A novel ultra-high-throughput 16S rRNA gene amplicon sequencing library preparation method for the Illumina HiSeq platform, Microbiome 5:68
- Hanan Abd El-Gawad (2016), Validation method of organochlorine pesticides residues in water using gas chromatography–quadruple mass, Water Science, Volume 30, Issue 2, October, Pages 96-107
- Handelsman (2004), Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiology and Molecular Biology Reviews. 2004; 68(4): 669-685. doi:10.1128/MMBR.68.4.669-685
- Jayanthi Abraham, Sivagnanam Silambarasan, Peter Logeswari, (2014) Simultaneous degradation of organophosphorus and organochlorine pesticides by bacterial consortium, Journal of the Taiwan Institute of Chemical Engineers, Volume 45, Issue 5, September, Pages 2590-2596
- Jayaraj, R., Megha, P. & Sreedev, P. (2017). Review Article. Organochlorine pesticides, their toxic effects on living organisms and their fate in the environment. Interdisciplinary Toxicology, 9(3-4), pp. 90-100.
- Loomis D, Guyton K, Grosse Y, El Ghissasi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Mattock H, Straif K (2015); Carcinogenicity of lindane, DDT, and 2,4-dichlorophenoxyacetic acid, Lancet Oncol, Aug;16(8):891-2.
- María S., Fuentes & Benimeli, Claudia & Cuozzo, Sergio & Saez, Juliana Maria & Amoroso, María. (2010). Microorganisms capable to degrade organochlorine pesticides. 1255-1264.
- Muir D, Sverko E (2006). Analytical methods for PCBs and organochlorine pesticides in environmental monitoring and surveillance: a critical appraisal. Analytical and Bioanalytical Chemistry, 386(4):769-789. doi:10.1007/s00216-006-0765-y.
- Pandey,P., P. S. Khillare, Krishan Kumar (2011), Assessment of Organochlorine Pesticide Residues in the Surface Sediments of River Yamuna in Delhi, India, Journal of Environmental Protection, 511-524
- Villa, S., Vighi, M., Maggi, V. (2003) Historical Trends of Organochlorine Pesticides in an Alpine Glacier Journal of Atmospheric Chemistry 46: 295.