Department of Life Science, Mount Carmel College, Autonomous, Bengaluru
Corresponding author email: sharadamma.n@mccblr.edu.in
Article Publishing History
Received: 06/07/2021
Accepted After Revision: 19/09/2021
Tannases or Tannin acyl hydrolases (EC 3.1.1.20) belonging to the superfamily of hydrolase finding their applications in food, brewing, chemical, and pharmaceutical industries etc., A number of fungal tannase were very well characterised. However, little is known about the extra cellular bacterial tannases and their properties. The present investigation was undertaken with an objective of tannic acid degradation from bacterial tannase. The bacterium was cultivated in MSM medium containing 0.3% tannic acid as sole carbon source and produced extracellular tannases from early lag phase (4 h) and increased exponentially till 10 h (Klebsiella pneumoniae BH49). The strain was confirmed by different biochemical tests and16S rDNA phylogenetic analysis. The crude enzyme showed maximum activity at pH 6.0 at 30 ºC and the activity was retained 99.33% at 40 ºC. The Km of the crude enzyme showed 0.0518 mM and 0.0389 mM for methyl gallate and propyl gallate as substrate respectively.
The specific activity of the crude enzyme was found to be 0.943U/mg for methyl gallate. In addition, the activity was significantly increased in presence of K+, Mg2+, and Ca2+ similarly, the activity was inhibited by Fe2+ Mn2+ and Cu2+. The enzyme was purified by ammonium sulphate fractionation followed by DEAE-Cellulose (Ion exchange chromatography). In gel staining confirmed major tannase enzyme and the SDS-PAGE analysis of the purified enzyme showed, the molecular weight of 55 kDa. Our investigations would give potential source for efficient production of extracellular tannase and can used for tannery effluent degradation, pharmaceutical and industrial applications.
Gallic Acid, Methyl Gallate, Propyl Gallate, Rhodanine.
Teron L, Rajgadia N, Dayana A T, Dhrithi P M, Nomuk K, Sharadamma N . Production of Tannin Acyl Hydrolase and its Purification from Klebsiella pneumoniae. Biosc.Biotech.Res.Comm. 2021;14(3)
Teron L, Rajgadia N, Dayana A. T, Dhrithi P. M, Nomuk K, Sharadamma N. Production of Tannin Acyl Hydrolase and its Purification from Klebsiella pneumoniae. Biosc.Biotech.Res.Comm. 2021;14(3). Available from: <a href=”https://bit.ly/38rUHDf“>https://bit.ly/38rUHDf</a>
Copyright © Teron et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Tannins are generally considered recalcitrant to biodegradation and major effluents of tanning industries and they pose potential threats to human health as well as the environment (Bari et al. 2015; Adamczyk et al. 2017). However, despite their toxic effects, some microorganisms have evolved to use gallotannins as carbon sources for growth by the action tannin acyl hydrolases, commonly known as tannases (Tannin acyl hydrolase E. C. 3.1.1.20).
Tannase, that hydrolysis of ester and depside bonds in varied substrates like gallotannins, gallic acid esters (Govindarajan et al. 2016a, Govindarajan et al. 2016b; Tripathi et al. 2018; Xu et al. 2019). Microorganisms are the main source of industrial enzymes due to their biochemical diversity and their amenability to genetic modifications. A significant number of tannase-producing microorganisms especially fungi and bacteria were identified (Thiyonila et al. 2020).
Fungal tannases have been well documented for their potential bioconversion and specifically for the biotransformation of tannic acid to gallic acid. To date, commercially available tannases are mainly produced by fungal species (Tripathi et al. 2016; Dhiman et al. 2018). However, the usage in industrial purpose is limited due to its lower catalytic efficiency in front of bacterial tannase.
Previous studies demonstrated that tannases from yeast have been only verified in Sporidiobolus ruineniae, Candida sp., Aureobasidium, Blastobotrys adeninivorans, and Kluyveromyces marxianus (Beniwal et al. 2010; Dhiman et al. 2018). Bacterial strains that belong to genera such as Staphylococcus, Lonepinella, Lactobacillus, Pseudomonas, Serratia, Bacillus, Azobacter, Klebsiella, Citrobacter, Pantonea, and Enterobacter were predominant (Tripathi et al. 2016; Thiyonila et al. 2020).
Meanwhile, the degradation of natural tannins by bacterial tannase were found to be very effective due to its industrial applications and its rapid catalytic degradation potential (Wang et al. 2019). It is crucial to isolate and identify the potential source for tannase-producing bacteria. In the present study, we have purified and biochemical properties were investigated for extracellular tannase from Klebsiella pneumoniae BH49 potential source for pharmaceutical and industrial applications.
MATERIAL AND METHODS
Tannic acid, Gallic acid, Methyl gallate, Propyl gallate, and Rhodanine were obtained from Sigma Chemical Co., St Louis, MO, USA. The culture media components used (Hi-Media, Mumbai), other chemicals used were of analytical grade. For the isolation of the microorganism, soil samples were collected from near Slaughter house habitat (Tannery Rd, Richards Town, Bengaluru, Karnataka, India).
About 1 gram of tannic acid was enriched to soil was serially diluted by suspending in 10 ml of sterile distilled water and the suspension was swirled about for a period of one to two hours. The serially diluted tubes (dilutions of 10-1, 10-2, 10-3, and 10-4) were plated on a LB Agar containing tannic acid. In total, four plates were made each plate containing a different concentration of tannic acid namely, 0.1%, 0.2%, 0.3% and 0.4% respectively.
For the identification of the microbial strain, morphological characteristics of the bacteria were determined by performing by microscopic observations, and the isolated pure strain was subjected to various biochemical tests according to Bergey’s manual of determinative bacteriology (Baird-Parker 1974). Further, the pure strain was confirmed by 16S rDNA and phylogenetic analysis (Sambrook and Russell 2001). For the analysis of substrates and products by thin layer chromatography and to determine the fate of the accumulated metabolites, the cells were pelleted by the configuration at 10,000 rpm for 10 min at 4 °C.
The supernatant containing gallic acid was extracted with ethyl ether (1:5 v/v), further the extracts were dried and resuspended in methanol. Thin layer chromatography was carried out by spotting the methanol extract with authentic substrate (tannic acid) and metabolite (gallic acid). The plates were developed using pentane saturated acetonitrile and toluene (2:1 v/v) containing 1.25% formic acid. The metabolites were visualized by exposing the plates to iodine vapors (Ferry et al. 1991).
For the production of extracellular Tannase by Klebsiella pneumoniae BH49 was analyzed in MSM supplemented with 0.3% tannic acid (filter sterilized). Extracellular tannase (Spent medium) was pooled in batch cultures in 100 ml Erlenmeyer flasks in three independent replicates. The crude enzyme at different time points were chilled and centrifuged at 10,000 rpm for 10 min at 4°C for further analysis. For enzyme assay, after implicating a few modifications, the activity of tannase was assayed by using methyl gallate as substrate. The activity was measured in terms of micromoles of gallic acid formation (Sharma et al. 2000).
For the biochemical characteristics of extracellular Tannase, temperature optima, the crude tannase activity was carried out by incubating enzyme and substrate at various temperatures ranging from 4°C to 100°C, the percentage activity and optimum temperature were determined spectroscopically. For temperature stability, enzyme stability was determined by incubating the enzyme along with the buffer for 20 min at different temperatures ranging from 4°C to 100°C. Further, the substrate was added and incubated at room temperature for an enzyme reaction.
For pH optima, the effect of pH on the enzyme activity was studied by conducting the reaction at various pH ranging from 3-10 and assayed for tannase to determine the pH optimum. For pH stability, enzyme stability was studied by pre-incubating the enzyme along with the buffer pH ranging from 3-10 for 20 min. Further, the substrate was added to the above solutions and incubated for 20 min. To determine the effect of metal ions on tannase activity, different metal like Fe2+, Zn2+, Ca2+, Mn2+, Mg2+, Co2+, K+ and Cu2+ were dissolved in citrate buffer (50 mM pH 6.0). The enzyme was preincubated in a buffer containing different metal ions for 15 min, and the reaction was initiated by adding substrate.
For the determination of Km and Vmax, different concentrations of methyl and propyl gallate were used for enzyme activity to determine Km and Vmax of the tannase was determined by Lineweaver-Burk plot. For the purification of extracellular Tannase, initially, the spent medium was subjected to ammonium sulfate fractionation (80%), followed by centrifugation for 12000 rpm for 15 min at 4 °C. The precipitated enzyme was resuspended and dialyzed in 10 mM citrate buffer pH 6.0 at 4 °C. The enzyme was further purified by DEAE-Cellulose column was equilibrated with 10 mM citrate buffer pH 6.0. The bound enzyme fractions were eluted (step-wise) by 200, 300, 500, and 700 mM NaCl in citrate buffer pH 6.0. Further, the enzyme fractions were pooled, dialyzed, and assayed for tannase.
Protein content was estimated by the Lowry method using bovine serum albumin as standard (Lowry et al. 1951). For In-gel staining of tannase activity, the electrophoresed gel (Native-PAGE) was incubated in methyl gallate (10 mM in 10 mM citrate buffer pH 6.0) for 30 min. The gel was washed with the same buffer and incubated in 0.667% methanolic rhodanine for 30 minutes at RT, further, 0.5 N KOH was added for color development. For the molecular weight determination by SDS-PAGE, the purified tannase was analyzed by SDS-PAGE (Laemmli 1970) and protein bands were stained by Coomassie Brilliant Blue R-250. The molecular weight of the enzyme protein was determined.
RESULTS AND DISCUSSION
Selection of bacterial strains: Eight strains of bacterial origin isolated from tannic acid-induced soil samples were evaluated for tannase. A total of 4 strains showed a colour change (dark colour) on LB agar plate with tannic acid after 48 h (Figure 1), were found to be positive tannase production and total activity was measured (Supplementary Figure 1). Based on the above observations Isolate C1 was selected for further studies. Similar studies were made for different bacterial strains (Sharma et al. 2000; Osawa et al. 2006, Murugan et al. 2007; Sivashanugam and Jayaraman 2011; Chartchai et al. 2020; Govindarajan et al. 2021).
Figure 1: Utilization of the Tannic Acid on LB Agar: Colour change (Dark) in LB agar plates containing 0.3% tannic acid.
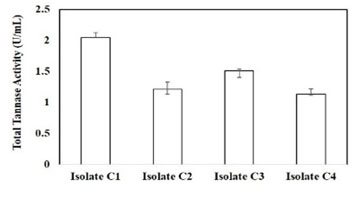
Supplementary Figure 1: Analysis of total tannase activity from different isolates
Characteristics of the Isolate C1 (potential) strain: Microscopic studies of Isolate C1 were found to be gram-negative cocci, non-motile and capsulated (Supplementary Figure 2 and Supplementary Table 1). Fermentation results revealed that dextrose, lactose, and sucrose were sole sources of carbon. The biochemical studies showed positive results for methyl red, Voges Proskauer, citrate, and production of catalase/oxidase (Table 1 and Supplementary Figure 3).
It has been observed that the optimal growth in MSM media with 0.3% tannic acid (pH 7.0) at 37°C. Based on the above observations and 16S rRNA studies revealed that the strain was found to be Klebsiella pneumoniae BH49 (Figure 2). Similar observations were also made for Klebsiella pneumoniae MTCC 7162 and Bacillus subtilis KMS2-2 (Sivashanmugam and Jayaraman 2011; Chartchai et al. 2020).
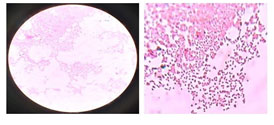
Supplementary Figure 2: Gram staining of Isolate C1- K. pneumoniae BH49
Supplementary Table 1: Colony Characteristics (Morphological Tests) of K. pneumoniae BH49 (Isolate C-1).
Table 1. Results of various biochemical tests carried out for tannic acid degrading bacteria K. pneumoniae BH49 (Isolate C-1)
| Colony | Morphological Observations |
| Size | 0.5 cm |
| Margin | Entire |
| Shape/form | Circular |
| Elevation | Unborate |
| Surface texture | Smooth |
| Consistency | Butter-ferous |
| Opacity | Trans-lucent |
| Chromo- genesis | Creamy white, off white |
| Biochemical test | Result |
| Lactose Fermentation | Positive |
| Dextrose Fermentation | Positive |
| Sucrose Fermentation | Positive |
| H2S Production | Negative |
| Citrate Utilization | Positive |
| Indole Test | Negative |
| MR Test (Methyl Red) | Positive |
| VP Test (Voges Proskauer) | Positive |
| Urease Activity | Negative |
| Catalase Activity | Positive |
| Oxidase Activity | Positive |
| Gelain Liquification | Negative |
| Starch Hydrolysis | Negative |

Supplementary Figure 3: Various Biochemical Tests of Tannic Acid Degrading Bacteria K. pneumoniae B (Isolate C-1).
Figure 2: Neighbor-joining tree showing the position of isolate Klebsiella pneumoniae BH49.

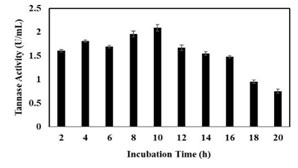
Production Tannase from K. pneumoniae BH49: It was observed that the bacteria can able to grow in tannic acid (0.3%) as a sole source of carbon. The results indicated that there was a significant increase in production of tannase at early lag phase (4 h) and the maximum degradation of tannic acid was observed in the exponential phase. The production rate was continuing till 10 h and was steadily decreased and the total enzyme activity was found to be the 2.14 U/mL of the crude extract (Figure 3).
Similar observations were made by group of bacterial strains of taxa Lactobacillus (0.3 U/ml), Bacillus sp. (1.03 U/ml), K. pneumoniae (0.75 U /ml), Enterobacter cloacae strain 41, Pseudomonas aeruginosa IIIB 8914 (13.65 and 12.90 U/ml) using amla and keekar leaves (2% w/v) respectively under SmF, K. pneumoniae MTCC 7162 (3.45 U/ml) and B. tequilensis K34.2 (0.60 U/mL) (Deschamps et al. 1983; Hadi et al. 1994; Kar et al. 2003; Selwal et al. 2010; Sivashanmugam and Jayaraman 2011; Govindarajan et al. 2019; Chartchai et al. 2020).
Figure 3: Analysis of Tannase activity at different time points.
Analysis of metabolites by Thin Layer Chromatography: The chromatogram revealed that the production of gallic acid, indicating that the hydrolysis was progressed with significant production of tannase. The Rf values gallic acid and pyrogallol were comparable with the Rf values of purified samples (Figure 4). Similar observations were made for the degradation of tannic acid in ruminal bacterium, Lactobacilli, Rhodococcus NCIM 2891, Enterococcus faecalis and Enterobacter cloacae (Nelson et al. 1995; Lee et al. 2008; Goel et al. 2011; Nadaf and Ghosh 2011; Govindarajan et al. 2019).
Figure 4: TLC profile of degradation of tannic acid: Lane 1-Standard tannic acid, Lane 2-Standard gallic acid, Lane 3-gallic acid and glucose.
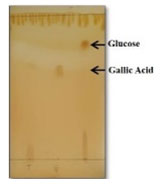
Properties of Extracellular Crude Tannase
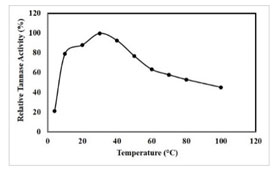
Determination of optimum temperature: The results revealed that the tannase was active in the temperature range of 4°C-100°C with an optimal activity (2.14 U/mL) at 40 °C (Figure 5), which was similar for K. pneumoniae MTCC 7162 at 40 °C, A. niger ATCC 16620, Bacillus licheniformis KBR 6 and for Bacillus subtilis KMS2-2 at 50 °C (Mondal and Pati 2000; Sabu et al. 2005; Sivashanmugam and Jayaraman 2011; Govindarajan et al. 2021).
Figure 5: Effect of temperature on tannase activity, results were obtained mean values of ± SD (n = 3).
Thermal stability of tannase: From the graph, it was revealed that the enzyme retained its activity above 99.5% at 40 °C (Figure 6). A similar result was observed for tannase produced from K. pneumoniae MTCC 7162 and Bacillus subtilis KMS2-2, which was thermally stable and retained its activity more than 50% at 60 °C and 80% at 50 °C (Sivashanmugam and Jayaraman 2011; Govindarajan et al. 2021). The obtained result suggests that the extracellular tannase from K. pneumoniae BH 49 persists its activity for a broad temperature range.
Figure 6: Effect of temperature stability of tannase activity, results were obtained mean values of ± SD (n = 3).
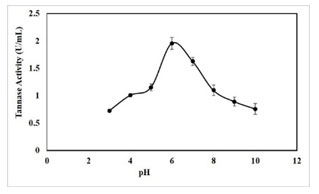
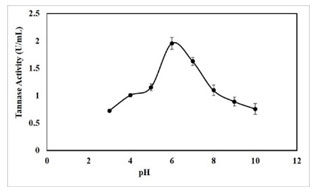
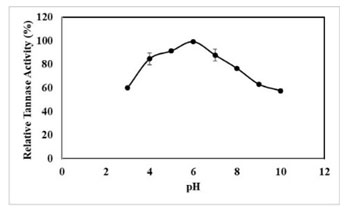
pH optima for tannase activity: The optimum pH for crude extract was found to be pH 6.0. It was observed that the enzyme showed a pH range of 3-10 with an optimal activity at pH 6.0 (1.96 U/mL) (Figure 7). Further, tannase activity was gradually decreased as pH reached the alkaline range. Optimum pH of 5.0-7.0 was reported for tannase from Paecilomyces variotii, pH 5.0 for P. chrysogenum, pH of 5.5 for K. pneumoniae MTCC 7162 and pH 6.0 for Bacillus subtilis KMS2-2 (Mahendran et al.
2006; Hamdy 2008; Sivashanmugam and Jayaraman 2011; Govindarajan et al. 2021). Some extracellular tannase from bacterial sources has been previously reported that the maximum activity at pH levels close to neutral (Mondal et al. 2001b; Batra and Saxena 2005; Sabu et al. 2006; Enemuor and Odibo 2009; Mahapatra et al. 2009; Chhokar et al. 2010).
Figure 7: Effect of pH on tannase activity, results were obtained mean values of ± SD (n = 3).
pH Stability of Tannase: The pH stability results of extracellular tannase revealed that the enzyme was stable from a pH range of 4-7 (Figure 8). Tannases from yeast and A. oryzae was stable in a wide range of pH (3.5-8.0), whereas tannases from P. chrysogenum and A. oryzae were stable in the narrow ranges of 4.5-6.0 and 5.0-5.5, respectively (Abdel‐Naby et al. 1999; Pan et al. 2020).
Stability of above 99.33% was retained by the crude enzyme at pH 6.0. Even at pH 4.0 and 8.0, the enzyme exhibited more than 70% of activity (Figure 8). Stability of above 81.3% and 80% was retained in tannase produced from K. pneumoniae MTCC 7162 at pH 6.0 and tannase from Acid Stable Yeast (Sivashanmugam and Jayaraman 2011; Kanpiengjai et al. 2021).
Figure 8: Effect of pH stability of tannase activity, results were obtained mean values of ± SD (n = 3).
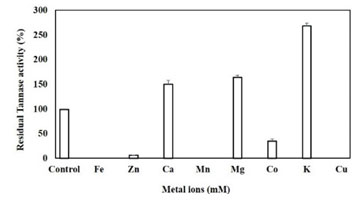
Effect of heavy metals: The tannase showed complete inhibition by Fe2+, Mn2+ and Cu2+, 95% by Zn2+ and 70% by Co2+ in 10 mM concentration. The obtained results and LB plots emphasis that the metal ions act as competitive inhibitors. However, the activity was almost doubled in presence of 10 mM K+, by 64% in presence of Mg2+ and up to 50% in presence of Ca2 (Figure 9). Previously it has been investigated that tannase from Aspergillus oryzae and P. chrysogenum was showed maximum inhibition in the presence of Fe2+, Zn2+, and Cu2+ (Ibuchi et al. 1968; Rajakumar and Nandy 1983). However, Tannase from A. niger was significantly inhibited by Cu2+ and to a lesser extent by Fe2+, Zn2+ (20 mM) (Barthomeuf et al. 1994).
Tannase from Bacillus subtilis KMS2-2 was inhibited by Cu2+ (40%), Fe2+ (56%), Fe3+ (31%), Ba2+ (39%), Hg2+ (48%) and K+ (32%), whereas, Zn2+ (08%), Ag2+ (23%) and Na+ (6%) showed moderate inhibitions. Among the metal ions studied, the Na+ significantly increased activity by 10% at 1 mM concentration; however, the activity was generally inhibited in the presence of large concentrations of ions, except Ca2+ ions. In the presence of Mg2+, the activity of tannase was enhanced (Kar et al. 2003; Chaitnyakumar and Anbalagan 2016; Govindarajan et al. 2021).
Figure 9: Effect of metals on Tannase activity, results were obtained mean values of ± SD (n=3).
Kinetic Parameters: Based on the the LB-Plot we obtained higher Km value for methyl gallate (0.0518 mM) and lower Km value for propyl gallate (0.0389 mM). However, the values of Km suggest Klebsiella pneumonia BH49 tannase showed more specific towards propyl gallate. The Km value is very low as compared to tannases from Selenomonas ruminantium and Cryphonectria parasatica using methyl gallate (1.6 mM and 7.49 mM respectively) (Farias et al. 1994).
The Km values for tannase produced by Rhodococcus NCIM 2891, Cryphonectria parasitica and Enterobacter cloacae (0.34 mM, 0.94 mM and 3.0 mM). Similarly, tannases from many fungi showed Km values of 0.20 to 1.03 mM for tannic acid (Farias et al. 1994; Ramirez-Coronel et al. 2003; Sabu et al. 2005; Mukerjee and Banerjee 2006; Nadaf and Ghosh 2011; Govindarajan et al. 2019).
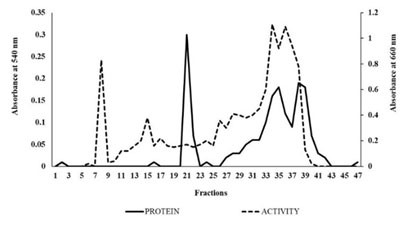
Purification of extracellular tannase: Tannase has been purified from Klebsiella pneumoniae BH49 to homogeneity by using the different steps (Table 2). The elution profile of extracellular tannase by DEAE-Cellulose showed the best yield of the enzyme (23%) (Figure 10). The specific activity of purified tannase was found to 0.943 U/mg for methyl gallate. Our results were compared to the purified tannase from Klebsiella pneumoniae MTCC 7162 and Bacillus subtilis KMS2-2 (Sivashanmugam and Jayaraman 2011; Govindarajan et al. 2021).
The yield of 7% was lower than the value reported for tannases from Penicillium chrysogenum and Cryphonectria parasitica (Farias et al. 1994; Rajakumar and Nandy 1983). The fold purification, on the other hand was identical to that of purified tannase from a variety of fungi and bacteria (Rajakumar and Nandy 1983; Sharma et al. 1999; Govindarajan et al. 2019).
Table 2. Purification table of tannase isolated from K. pneumoniae BH49
| Purification steps | Total volume (mL) | Total activity (U) | Total Protein (mg) | Specific activity (U/mg) | Yield (%) | Purification fold |
| Crude enzyme | 20 | 41.8 | 112 | 0.373 | 100 | 1 |
| Ammonium sulfate precipitation (0-80%) | 10 | 27.5 | 85 | 0.323 | 65.78 | 0.865 |
| DEAE – Cellulose | 10 | 9.84 | 10.45 | 0.941 | 23.54 | 2.522 |
Figure 10: The elution profile of DEAE-Cellulose Ion exchange chromatography of Tannase: Protein fractions (-) and Enzyme fractions (—).
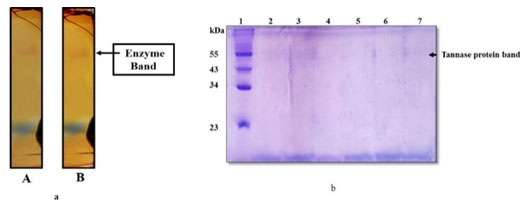
In-gel staining of tannase activity: Tannase activity of both crude and purified enzyme (In-gel) was visualized by formation of chromogen by methanolic rhodanine in KOH, the chromogenic bands that were obtained (Figure 11a).
Molecular weight determination of Extracellular purified Tannase: The SDS-PAGE profile of purified tannase showed one major band with the molecular mass of about 55 kDa (Figure 11b and Supplementary Figure 4). It was shown that most of the purified fungal tannases range from168 to 310 kDa and multimeric (Aguilar 2001; Ramirez 2003). Similarly, it has been reported the isoforms of tannase from Paecilomyces variotii of 87.3 kDa and 71.5 kDa (Battstin and Macedo 2007).
Tannase from Verticillium sp. P9 had two subunits with molecular masses of 39.9 and 45.6 kDa (Kasieczka 2007). In addition, the molecular mass of tannase from K. pneumoniae MTCC 7162 and Bacillus subtilis KMS2-2 by SDS-PAGE revealed that 46.5 kDa and 43 kDa respectively (Sivashnmugam and Jayaraman 2011; Govindarajan et al. 2021).
Figure 11: a, Native-PAGE pattern of Tannase activity; A-Crude enzyme, B-Purified tannase. b, SDS-PAGE profile of purified tannase; Lane1-Molecular weight marker, Lane-2 to 7 purified tannase.
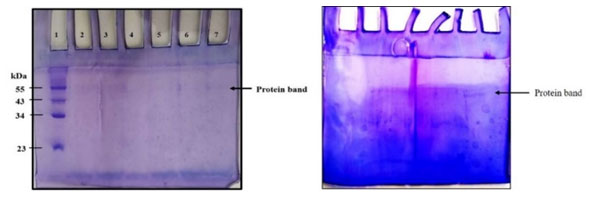
Supplementary Figure-4: SDS-PAGE profile of purified tannase; Lane1-Molecular weight marker, Lane-2 to 7 purified tannase.
CONCLUSION
The findings of the present study explored the production of tannase from Klebsiella pneumonia BH49 has shown a higher affinity for tannic acid compared to other bacterial tannases. The bacteria produced a high amount of extracellular tannase in the exponential phase to be a conventional strain compared to fungal strains. The biochemical properties like temperature stability, pH stability and other kinetic parameters further insights to potential source for efficient production of extracellular tannase and can be used for tannery effluent degradation, pharmaceutical and industrial applications. In addition, the production of gallic acid (antioxidant) was another significant end product and could the novel discovery used to remove tannins from food fodder and thus improve the health of the animals.
ACKNOWLEDGEMENTS
This study was financially supported by management (fund provided for Post graduate student research projects). Also, authors would like to thank Dr. Suba G Manuel, Associate Professor, Department of Life Science, Mount Carmel College, Autonomous, Bengaluru. India, for the support and guidance.
Conflicts of Interests: Authors declare no conflicts of interests to disclose.
Author Contributions: Sharadamma N designed the study, analysed the results, and prepared the manuscript. All authors performed the experiments and analysed the results, reviewed the results and approved the final version of the manuscript.
REFERENCES
Abdel‐Naby, M.A., Sherif, A.A., El‐Tanash, A.B. et al. (1999). Immobilization of Aspergillus oryzae tannase and properties of the immobilized enzyme. Journal of Applied Microbiology, 87(1), pp.108-114.
Aguilar, C. N., and Gutiérrez-Sánchez, G (2001). Sources, properties, applications and potential uses of tannin acyl hydrolase. Food Science and Technology International, 7 (5), 373-382.
Adamczyk, B., Simon, J., Kitunen, V., et al. (2017). Tannins and their complex interaction with different organic nitrogen compounds and enzymes: old paradigms versus recent advances. Chemistry Open, 6(5), p.610.
Baird-Parker, A. C., Buchanan, R. E., and Gibbons, N. E (1974). Bergey’s manual of determinative bacteriology. The Williams and Wilkins Company, Baltimore, 478-489.
Bari, M.L., Simol, H.A., Khandoker, N., et al. (2015). Potential human health risks of tannery waste-contaminated poultry feed. Journal of Health and Pollution, 5(9), pp.68-77.
Barthomeuf, C., Regerat, F., and Pourrat, H (1994). Production, purification and characterization of a tannase from Aspergillus niger LCF 8. Journal of Fermentation and Bioengineering, 77 (3), 320-323.
Batra, A., and Saxena, R. K (2005). Potential tannase producers from the genera Aspergillus and Penicillium. Process Biochemistry, 40(5), 1553-1557.
Battestin, V., and Macedo, G. A (2007). Purification and biochemical characterization of tannase from a newly isolated strain of Paecilomyces variotii. Food Biotechnology, 21(3), 207-216.
Beniwal, V., and Chhokar, V (2010). Statistical optimization of culture conditions for tannase production by Aspergillus awamori MTCC 9299 under submerged fermentation. Asian J Biotechnol, 2 (1), 46-52.
Chaitanyakumar, A., and Anbalagan, M (2016). Expression, purification and immobilization of tannase from Staphylococcus lugdunensis MTCC 3614. AMB Express, 6 (1), 1-9.
Chhokar, V., Sangwan, M., Beniwal, V., et al. (2010). Effect of additives on the activity of tannase from Aspergillus awamori MTCC9299. Applied Biochemistry and Biotechnology, 160 (8), 2256-2264.
Dhiman, S., Mukherjee, G., and Singh, A. K (2018). Recent trends and advancements in microbial tannase-catalyzed biotransformation of tannins: a review. International Microbiology, 21(4), 175-195.
Enemuor, S. C., and Odibo, F. J. C (2009). Culture conditions for the production of a tannase of Aspergillus tamarii IMI388810 (B). African Journal of Biotechnology, 8 (11).
Farias, G. M., Gorbea, C., Elkins, J. R., et al. (1994). Purification, characterization, and substrate relationships of the tannase from Cryphonectria parasitica. Physiological and Molecular Plant Pathology, 44 (1), 51-63.
Ferry, J., and Larson, R. A (1991). A mixed solvent for rapid TLC analysis of phenolic compounds. Journal of chromatographic science, 29 (11), 476-477.
Goel, G., Kumar, A., Beniwal, V., et al. (2011). Degradation of tannic acid and purification and characterization of tannase from Enterococcus faecalis. International biodeterioration & biodegradation, 65 (7), 1061-1065.
Govindarajan, R.K., Revathi, S., Rameshkumar, N., et al. (2016a). Microbial tannase: current perspectives and biotechnological advances. Biocatal. Agric. Biotechnol. 6, 168-175.
Govindarajan, R.K., Revathi, S., Rameshkumar, N., et al. (2016b). Isolation and Characterization of Tannase Producing Bacteria from the gut of Gryllotalpa krishnani. J. Microbiol. Biotech. Food Sci. 6, 813-817.
Govindarajan, R.K., Krishnamurthy, M., Neelamegam, R., et al. (2019). Purification, structural characterization and biotechnological potential of tannase enzyme produced by Enterobacter cloacae strain 41. Process Biochemistry, 77, pp.37-47.
Govindarajan, R.K., Mathivanan, K., Khanongnuch, C., et al. (2021). Tannin acyl-hydrolase production by Bacillus subtilis KMS2-2: Purification, characterization, and cytotoxicity studies. Journal of King Saud University-Science, 33(3), p.101359.
Hadi, T. A., Banerjee, R., and Bhattacharyya, B. C (1994). Optimization of tannase biosynthesis by a newly isolated Rhizopus oryzae. Bioprocess Engineering, 11(6), 239-243.
Kar, B., Banerjee, R., and Bhattacharyya, B. C (2003). Effect of additives on the behavioural properties of tannin acyl hydrolase. Process Biochemistry, 38 (9), 1285-1293.
Kanpiengjai, A., Khanongnuch, C., Lumyong, S., et al. (2020). Co-production of gallic acid and a novel cell-associated tannase by a pigment-producing yeast, Sporidiobolus ruineniae A45. 2. Microbial cell factories, 19(1), pp.1-12.
Kasieczka-Burnecka, M., Kuc, K., Kalinowska, H., et al. (2007). Purification and characterization of two cold-adapted extracellular tannin acyl hydrolases from an Antarctic strain Verticillium sp. P9. Applied Microbiology and Biotechnology, 77 (1), 77-89.
Kwon, T.Y., Shim, S.M. and Lee, J.H (2008). Characterization of lactobacilli with tannase activity isolated from Kimchi. Food science and biotechnology, 17(6), pp.1322-1326.
Laemmli, U. K (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680-685.
Lowry, O. H., Rosebrough, N. J., Farr, A. L., et al. (1951) Protein measurement with the Folin phenol reagent. Journal of biological chemistry, 193, 265-275.
Mahapatra, S., and Banerjee, D (2009). Extracellular tannase production by endophytic Hyalopus sp. The Journal of general and applied microbiology, 55(3), 255-259.
Mahendran, B., Raman, N., and Kim, D. J (2006). Purification and characterization of tannase from Paecilomyces variotii: hydrolysis of tannic acid using immobilized tannase. Applied Microbiology and Biotechnology, 70 (4), 444-450.
Mondal, K. C., and Pati, B. R (2000). Studies on the extracellular tannase from newly isolated Bacillus licheniformis KBR 6. Journal of Basic Microbiology: An International Journal on Biochemistry, Physiology, Genetics, Morphology, and Ecology of Microorganisms, 40 (4), 223-232.
Mondal, K. C., Banerjee, D., Banerjee, R., et al. (2001b). Production and characterization of tannase from Bacillus cereus KBR9. The Journal of general and applied microbiology, 47 (5), 263-267.
Mukherjee, G., and Banerjee, R (2006). Effects of temperature, pH and additives on the activity of tannase produced by a co-culture of Rhizopus oryzae and Aspergillus foetidus. World Journal of Microbiology and Biotechnology, 22 (3), 207-212.
Murugan, K., Saravanababu, S., and Arunachalam, M (2007). Screening of tannin acyl hydrolase (EC 3.1. 1.20) producing tannery effluent fungal isolates using simple agar plate and SmF process. Bioresource Technology, 98 (4), 946-949.
Nadaf, N. H., and Ghosh, J. S (2011). Production, purification and characterization of tannase from Rhodococcus NCIM, 2891. Current Research Journal of Biological Sciences, 3 (3), 246-253.
Nelson, K. E., Pell, A. N., Schofield, P., et al. (1995). Isolation and characterization of an anaerobic ruminal bacterium capable of degrading hydrolysable tannins. Applied and environmental microbiology, 61(9), 3293.
Osawa, R., Fujisawa, T., and Pukall, R (2006). Lactobacillus apodemi sp. nov., a tannase-producing species isolated from wild mouse faeces. International journal of systematic and evolutionary microbiology, 56 (7), 1693-1696.
Pan, J., Wang, N.N., Yin, X.J., et al. (2020). Characterization of a Robust and pH-Stable Tannase from Mangrove-Derived Yeast Rhodosporidium diobovatum Q95. Marine Drugs, 18(11), p.546.
Ramírez-Coronel, M. A., Viniegra-Gonzalez, G., Darvill, A., et al. (2003). A novel tannase from Aspergillus niger with β-glucosidase activity. Microbiology, 149 (10), 2941-2946.
Sabu, A., Augur, C., Swati, C., et al. (2006). Tannase production by Lactobacillus sp. ASR-S1 under solid-state fermentation. Process Biochemistry, 41(3), 575-580.
Sambrook, J. and Russell, D.W (2001). Molecular cloning. A Laboratory Manual. Third edition Cold Spring Harbor Laboratory Press Cold Spring Harbor NY, 17-18.
Selwal, M. K., Yadav, A., Selwal, K. K., et al. (2010). Optimization of cultural conditions for tannase production by Pseudomonas aeruginosa IIIB 8914 under submerged fermentation. World Journal of Microbiology and Biotechnology, 26 (4), 599-605.
Sharma, S., Bhat, T. K., and Dawra, R. K (2000). A spectrophotometric method for assay of tannase using rhodanine. Analytical Biochemistry, 279 (1), 85-89.
Sivashanmugam, K., and Jayaraman, G (2011). Production and partial purification of extracellular tannase by Klebsiella pneumoniae MTCC 7162 isolated from tannery effluent. African Journal of Biotechnology, 10 (8), 1364-1374.
Kwon, T., Shim S., and Lee, J.H. (2008). Characterization of Lactobacilli with tannase activity isolated from kimchi. Food Science and Biotechnology, 17 (6), 1322-1326.
Thiyonila, B., Kannan, M., Reneeta, N.P., et al. (2020). Influence of tannase from Serratia marcescens strain IMBL5 on enhancing antioxidant 2 properties of green tea. BCAB-101675. Biocat. Agric. Biotechnol.
Tripathi, A. D., and Sharma, K. B. L (2016). Study on tannase producing Bacillus megaterium isolated from tannery effluent. Int. J. Adv. Res. Biol. Sci, 3 (7), 28-35.
Tripathi, A.D. and Lakshmi, B (2018). Statistical optimization of extracellular tannase production by Streptomyces sp. AT 13 using response surface methodology and Plackett-Burmen design. Bioscience Biotechnology Research Communications, 11(4), pp.691-698.
Unban, K., Kodchasee, P., Shetty, K. et al. (2020). Tannin-tolerant and extracellular tannase producing Bacillus isolated from traditional fermented tea leaves and their probiotic functional properties. Foods, 9(4), p.490.
Wang, D., Liu, Y., Lv, D., et al. (2019). Substrates specificity of tannase from Streptomyces sviceus and Lactobacillus plantarum. International Microbiology 21, 175-195.
Xu, X.Y., Meng, J.M., Mao, Q.Q., et al. (2019). Effects of Tannase and Ultrasound Treatment on the Bioactive Compounds and Antioxidant Activity of Green Tea Extract. Antioxidants 8, 362.


