KSV-Center for Skill and Research Enhancement, Kadi Sarva
Vishwavidyalaya, Gandhinagar, 382015, India
Corresponding author email: bjarullah@gmail.com
Article Publishing History
Received: 12/12/2022
Accepted After Revision: 25/02/2023
With an increasing rate of global warming and unstable climatic conditions concerns with regards to epidemiology of plant viruses are on the rise. Studies suggest accelerating climatic changes shall severely affect the management of pest and diseases in cultivated crops. Tomato Leaf Curl Virus (TLCV) is an economically affecting viral infections of tomato (Lycopersicum esculentum). The disease causes severe yield loss and major economic impairment. The current study was therefore taken up to understand the influence of agro climatic zones on diversity of TLCV in Tomato plants. Samples of TLCV infected Tomato plants exhibiting varying symptoms were collected from seven different agro climatic zones of Gujarat followed by isolation of viral particles, molecular characterization and development of phylogenetic tree. Interestingly the molecular analysis of the isolated viral samples indicated little influence of climatic conditions on the types of TLCV infecting the tomato plants.
Begomovirus, Lycopersicum esculentum, Gujarat, Solanaceous, TLCV.
Golden T. P, Jarullah B. Phylogeny of strains of Tomato Leaf Curl Virus from Agroclimatic Zones of Gujarat. Biosc.Biotech.Res.Comm. 2023;16(1).
Golden T. P, Jarullah B. Phylogeny of strains of Tomato Leaf Curl Virus from Agroclimatic Zones of Gujarat. Biosc.Biotech.Res.Comm. 2023;16(1). Available from: <a href=”https://bit.ly/3XBljJd“>https://bit.ly/3XBljJd</a>
INTRODUCTION
Tomato (Lycopersicum esculentum) is an essential solanaceous crop cultivated worldwide. After China, India is the highest tomato producer of the world. Gujarat has a second highest productivity after Karnataka. Viral infections are the major cause of economically devastating diseases in tomato. Of these Begomovirus is the most widespread and deeply studied genera of plant virus comprising 162 known species infecting tomato. Begomoviruses are monopartite or bipartite, whitefly-transmitted geminiviruses that are found in the Eastern (both genome types) and Western Hemisphere (only bipartite are thought to be endemic. The acceleration in tomato virus discovery is far superior to the post discovery characterization. This lag leads to continued economic damages from known virus as well. (Shankar 2021, Rivarez et al. 2021, Bozbuga et al 2022).
Tomato Leaf Curl Virus (TLCV) is one such economically affecting viral infections of tomato. The disease causes symptoms such as leaf lamina yellowing, upward curling and distortion of leaf, decrease in size of new leaves, wrinkled appearance of leaf, decrease in internodes, reduction in height of plants, and flower drop from plant prior to fruiting. All this occurs within 2 to 3 weeks after infection. The disease was first identified in Israel. In Indian subcontinent, Tomato Leaf Curl Disease (TLCD) is a plays a major role in reduced crop productivity for tomato cultivators with numerous reports on new strains being documented (Rivarez et al. 2021). Tomato Leaf Curl Virus (TLCV) is a viral disease, transmitted by vector whitefly. TLCV belongs to geminivirade family and has a small geminate particle of 20×30 nm.
Genome of TLCV consists of both mono- and bipartite genomes encapsulated by single capsid protein. Although some Indian TLCV isolates such as tomato leaf curl New Delhi virus (TLCNDV) and tomato leaf curl Palampur virus (TLCPalV) are bipartite (DNA-A and DNA-B) in nature, most of the TLCV isolates reported to date have monopartite (DNA-A) genome organization. These are single stranded DNA genomes having a size of approximately 2.7 kb size. They code for viral factors needed for viral replication, transmission, encapsidation and spread (Reddy et al. 2005; Wendy et.al. 2020, Sohrab et. al. 2021).
Current global environmental concerns such as unpredictable climatic conditions and global warming increases probabilities of poor management of pest and diseases in the cultivated crops (Trebicki P. 2020). In order to facilitate virus disease forecasting and prevention of viral disease outbreak in tomato it is important to understand the epidemiology of these viruses. The current study therefore focuses on the molecular characterization of TLCV to understand the influence of agro climatic zones on its diversity in Tomato plants.
MATERIALS & METHODS
Sample collection: Samples of TLCV infected Tomato plants exhibiting varying symptoms were collected from seven different agro climatic zones of Gujarat (Indian Horticulture Database., 2011). The virus were classified into 11 strains based on the symptoms and geographical location of collection site.
Isolation of viral samples: The collected leaf samples were washed with water and were stored in -20oC deep freezer. 100 grams of leaves were homogenized using mortar and pestle in 0.1 M Phosphate buffer. The suspension was then filtered through muslin cloth. The filtrate was collected and stirred at 4oC with half volume of Chloroform. After 30 minutes the mixture was centrifuged at 10000 rpm for 15 minutes. After centrifugation, PEG and NaCl were added to the supernatant while stirring at 4oC. After 2 hours for 60 minutes centrifugation of the mixture was done at 16000 rpm. The precipitates were dissolved in 100μl of 0.1 M Potassium Phosphate buffer, pH 7.8. This was filtered using 0.45μ Syringe filter (Palmer et al.,1998).
To confirm the isolation of viral particles, the samples were characterized by Transmission Electron Microscopy (TEM).
DNA extraction: DNA extraction was done for the isolated viral samples using TempliPhi kit, GE Healthcare, U.S.A. Rolling circle amplification (RCA) was used to amplify the DNA. The obtained DNA was digested using Kpn I. The resultant fragment (∼2.8-kb) was cloned in pUC18 plasmid. The full genome sequence was determined for all the viral samples obtained.
Phylogenetic Analysis: The nucleotide sequences obtained in this study were deposited in GenBank. Sequences were retrieved and a phylogenetic tree of all the 11 samples along with one outgroup of TLCV retrieved from NCBI database was constructed using CLUSTAL W program (random seed number, 111; bootstrap value, 1000).
RESULTS AND DISCUSSION
Collection of samples from different agroclimatic regions: Twenty six samples of tomato leaves infected with symptoms similar to TLCV were collected from different agroclimatic regions of Gujarat. The viral samples were classified into 11 strains based on the symptoms observed and geographical location of collection site (Table I).
Table 1: Types of virus found in different agro climatic zones
| Code | Agro climatic zones | Sample collection region |
| GJ 1 | South Gujarat Heavy rainfall Zone | Valsad |
| GJ 2 | South Gujarat Zone | Surat, Ankleshwar |
| GJ 3 | Middle Gujarat Zone | Balashinor, Matar, Kheda |
| GJ 4 | North-West Zone | Vijapur, Himatnagar |
| GJ 5 | North Saurashtra Zone | Botad |
| GJ 6 | South Saurashtra Zone | Kodinar, Jetpur |
| GJ 7 | Bhal and Costal Zone | Dholka |
Isolation of viral samples:
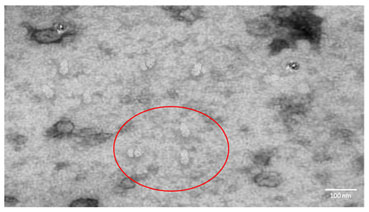
The virus were isolated and confirmed to be TLCV by TEM (Figure I).
Figure 1: Electron Microscopic image of the Tomato Leaf Curl Viral particles
Molecular Analysis: Total 11 samples were sent for molecular analysis and the sequencing was done by a third party institution. The sequences were submitted and the Genbank ID was procured. The details were given in the Table II.
Table 2. GenBank Accession Numbers for sequenced genomes
| Sr. No. | Area | GenBank Accession Number | ||
| DNA-A | DNA-B | |||
| 1 | Ankleshwar | MT551610 | MT316381 | MT295294 |
| 2 | Balashinor | MT551611 | MT316382 | MT295295 |
| 3 | Botad | MT551612 | MT316383 | MT295296 |
| 4 | Dholka | MT551613 | MT316384 | MT295297 |
| 5 | Himatnagar | MT551614 | MT316385 | MT295298 |
| 6 | Jetpur | MT551615 | MT316386 | MT295299 |
| 7 | Kodinar | MT551616 | MT316387 | MT295300 |
| 8 | Matar | MT551617 | MT316388 | MT295301 |
| 9 | Surat | MT551618 | MT316389 | MT295302 |
| 10 | Valsad | MT551620 | MT316390 | MT263149 |
| 11 | Vijapur | MT551621 | MT316391 | MT295303 |
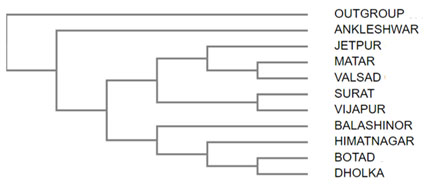
Phylogenetic Tree: The phylogenetic tree was developed from the molecular study of the different samples of TLCV collected from the different agro climatic zones of Gujarat (Figure II).
Figure 2: Cladogram of the obtained genomic sequences
DISCUSSION
The need to surpass food production over population explosion not only demands increasing production but also decreasing yield loss (Jones et.al. 2012). During the current times of climatic instability, studying the influence of climatic alterations on the prevalence of disease in wild type and cultivated plants is therefore of paramount importance. Viruses cause almost half of the evolving plant diseases worldwide (Jones 2019). Despite all investigations in viral diseases control, no antiviral products are available for plant disease management till now (Petrov et.al. 2021). In the past decade 45 novel species of virus infecting tomato have been identified (Rivarez et al. 2021). Tomato leaf curl disease (TLCD) is the most common viral disease in the tomato plant (Arooj et.al. 2017). In order to facilitate virus disease forecasting and enhance TLCV management, it is vital to understand the epidemiology of the disease. The current study therefore focused on, the molecular characterization of TLCV from different agro climatic zones of Gujarat with special attention on understanding the climatic influence on its diversity in Tomato plants.
Several previous reports suggest temperature to affect susceptibility and symptom development of other viral diseases ( Llamas-Llamas et. al. 1998; Zhang et. al. 2012; Prasch et. al. 2013; Zhong et. al. 2013; Ghoshal et. al. 2014; Patil et. al. 2015). These studies indicate the one type of infection under specific climatic conditions should predominate other infections. From our results it was interesting to observe that the sequences of viruses obtained from the same agro climatic zones were genetically very different. The viruses were separated into two main clads one consisted of Dholka, Botad, Himatnagar and Balashinor. Each of these isolates were from different agro climatic zone. The second clad had viruses from all agro climatic zones. No distinct pattern of viral infection based on agroclimatic zone was observed. Virus isolated from infected plants of Ankleshwar, South Gujarat Zone showed totally distinct genetic makeup.
Although previously we have reported severity of TLCV infection to influenced by climatic factors (Shelat et al.,2014), the current study confirms the type of TLCV strain not to be influenced by the climatic conditions. We therefore conclude little influence of climatic parameters on the types of TLCV strain infecting the tomato plants.
Conflict of interest:No conflict of interests
Data Availability Statement: All data / results / information is available with the authors and can be shared on a reasonable request made to the corresponding author when required.
REFERENCES
Amari, K., Huang, C. & Heinlein, M., 2021. Potential Impact of Global Warming on Virus Propagation in Infected Plants and Agricultural Productivity. Frontiers in Plant Science, March.Volume 12.
Anon., 2011. Indian Horticulture Database
Arooj, S. et al., 2017. Management Of Tomato Leaf Curl Virus Through Non-Chemicals In Relation To Environmental Factors. Pakistan Journal of Phytopathology, July, Volume 29, p. 41.
Bozbuga, R. et al., 2022. Host-Pathogen and Pest Interactions: Virus, Nematode, Viroid, Bacteria, and Pests in Tomato Cultivation. In: Tomato – From Cultivation to Processing Technology
Ghoshal, B. & Sanfaçon, H., 2014. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology, May, Volume 456-457, p. 188–197.
Jones, R. A. C. & Barbetti, M. J., 2012. Influence of climate change on plant disease infections and epidemics caused by viruses and bacteria.. CABI Reviews, February, Volume 2012, p. 1–33.
Jones, R. A. C. & Naidu, R. A., 2019. Global Dimensions of Plant Virus Diseases: Current Status and Future Perspectives. Annual Review of Virology, September, Volume 6, p. 387–409.
Llamas-Llamas, M. E. et al., 1998. Effect of temperature on symptom expression and accumulation of tomato spotted wilt virus in different host species. Plant Pathology, June, Volume 47, p. 341–347.
Palmer, K. E., Schnippenkoetter, W. H. & Rybicki, E. P., 1998. Geminivirus Isolation and DNA Extraction. In: Plant Virology Protocols. Humana Press, p. 41–52.
Patil, B. L. & Fauquet, C. M., 2014. Light intensity and temperature affect systemic spread of silencing signal in transient agroinfiltration studies. Molecular Plant Pathology, October, Volume 16, p. 484–494.
Petrov, N. &. S. & Mariya & Gaur, R., 2021. Plant Virus-Host Interaction. In: T. H. R.K. Gaur & P. Sharma, eds. Elsevier, p. 469–488.
Prasch, C. M. & Sonnewald, U., 2013. Simultaneous Application of Heat, Drought, and Virus to Arabidopsis Plants Reveals Significant Shifts in Signaling Networks. Plant Physiology, June, Volume 162, p. 1849–1866.
Reddy, R. V. C., Colvin, J., Muniyappa, V. & Seal, S., 2005. Diversity and distribution of begomoviruses infecting tomato in India. Archives of Virology, February, Volume 150, p. 845–867.
Rivarez, M. P. S. et al., 2021. Global Advances in Tomato Virome Research: Current Status and the Impact of High-Throughput Sequencing. Frontiers in Microbiology, May.Volume 12.
Rivarez, M. P. S. et al., 2021. Global Advances in Tomato Virome Research: Current Status and the Impact of High-Throughput Sequencing. Frontiers in Microbiology, May.Volume 12.
Shankar, A., 2021. Tomato Leaf Curl Virus of Tomato: Scenarios and Confronts. August.
Shelat, M. et al., 2014. Prevalence and distribution of Tomato leaf curl virusin major agroclimatic zones of Gujarat. Advances in Bioscience and Biotechnology, Volume 05, p. 1–3.
Trebicki, P., 2020. Climate change and plant virus epidemiology. Virus Research, September, Volume 286, p. 198059.
Zhang, X. et al., 2012. Temperature-Dependent Survival of Turnip Crinkle Virus-Infected Arabidopsis Plants Relies on an RNA Silencing-Based Defense That Requires DCL2, AGO2, and HEN1. Journal of Virology, June, Volume 86, p. 6847–6854.
Zhong, S.-H.et al., 2013. Warm temperatures induce transgenerational epigenetic release of {RNA} silencing by inhibiting {siRNA} biogenesis in Arabidopsis. Proceedings of the National Academy of Sciences, May, Volume 110, p. 9171–9176.


