Department of Biotechnology and Agricultural Extension, M. S. Swaminathan School of Agriculture, Centurion University of Technology and Management, Parlakhemundi, Odisha 761211 India
Corresponding author Email: preetha.bhadra@gmail.com
Article Publishing History
Received: 10/07/2019
Accepted After Revision: 20/09/2019
Genetically changed or designed foods are made from speedily increasing technologies that have sparked international debates and considerations regarding health and safety. These considerations target the potential dangers to human health, the risks of genetic pollution, and also the death of other farming techniques additionally as theft and economic exploitation by massive non-public firms. Transgenic or genetically changed plants possess novel genes that impart useful characteristics like weed killer resistance. One amongst the smallest amount understood areas within the environmental risk assessment of genetically changed crops is their impact on soil- and plant-associated microorganism communities. The popularity that these interactions might amendment microorganism multifariousness and have an effect on scheme functioning has initiated a restricted range of studies within the space. Moreover, novel proteins are shown to be free from transgenic plants into the soil scheme, and their presence will influence the multifariousness of the microorganism community by selection stimulating the expansion of organisms that may use them. Microorganism diversity is altered once related to transgenic plants; but these effects are each variable and transient. Minor alterations within the diversity of the microorganism community might have an effect on soil health and scheme functioning, and so, the impact that plant selection might wear the dynamics of the rhizosphere microorganism populations and successively plant growth and health and scheme property need additional study. Our aim is to identify the genes by the multiple sequence alignment (MSA) or the proteins related to the gene which are causing health hazard in human and to reduce the risk by homology modeling and spectroscopic analysis
Genetically modified crops, Homology Modeling, Multiple Sequence Alignment, Spectroscopic Analysis, Trans-genes
Bhadra P, Deb A. On the Reduction of Health Hazards Caused by Modified Genes in Indigenous Rice Plant Varieties. Biosc.Biotech.Res.Comm. 2019;12(3).
Bhadra P, Deb A. On the Reduction of Health Hazards Caused by Modified Genes in Indigenous Rice Plant Varieties. Biosc.Biotech.Res.Comm. 2019;12(3). Available from: https://bit.ly/2kUKOZL
Copyright © Bhadra and Deb, This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
Ranchers have with progress turned to hereditarily rising their yields through pondered plant rearing for a huge number of years, however, its logical premise wasn’t set up till established botanist hereditary science were rediscovered inside the mid-twentieth century. For centuries, the typical plant reproducing practices of ranchers have the diode to fixing the cosmetics and advancement of harvests. amid this sense, ranchers are thought of to be the essential hereditary designers (Dunfield et al. 2004) desoxyribonucleic (DN) corrosive sequencing from qualities (Walters ,2004) exploitation DN corrosive markers (Acosta and Chaparro, 2008) for building hereditary maps, concocting PCR-based procedures for picking and portraying qualities, and DN corrosive exchange advancements between totally unique species have all set the establishments for the popular generation of the hereditarily designed plants and yields by and by available (Fischer and Emans,2000; Uzogara, 2000; Margulis,2006; Singh et.al. 2006).
The apprehensions of the general population contradicting the innovation creating GM crops are related with a more extensive range of issues including the inclination that transnational organizations are more keen on expanding their benefits than in ensuring the earth or easing hunger, the likelihood that transgenic harvests may attack wild biological systems with hindering consequences for biodiversity, the unjustifiable challenge with other rural frameworks, for example, natural, agro-environmental and customary ones, the negative impacts that GM sustenance may deliver on human wellbeing,(ILSI,2003 and 2004; Halpin ,2005; Brookes et.al,2006; ISAAA ,2006; Made et.al,2006;Taylor et.al, 2006; Jeong et al.,2007;) the conceivable negative effect of GM crops on nourishment supply security, and an absence of trust in the offices in charge of directing GM crop bio-security. Commentators of GM crops contend that transgenic innovation has genuine ramifications for ranchers in creating nations as this remote global innovation may crush ranchers’ abilities worked around indigenous agrarian frameworks, subsequently intensifying social rejection on account of subsistence ranchers (Chakraborty et.al,2001; WAL ,2001; Bucchin and Goldman,2002; Newell-Mcgloughlin,2006; Alexander et.al,2007; Oliver et.al, 2007; Raney and Pingali, 2007).
As per some examination, numerous anti-microbial gatherings have been advancing neoliberal rationale as they have concentrated on the contemporary markets as the most ideal method for guaranteeing elective agribusiness. Their strategies stay concentrated on what should be possible at the ware level (Hodgson, 2001; Setamou et.al. 2002; Tolstrup et.al, 2003; Munoz et. Al, 2004; Poulsen et.al, 2007; Schrøder et.al. 2007).
The original GM crops designed with info qualities to give improved agronomic execution and monetary and natural advantages have been related to ranchers’ interests. Be that as it may, there is an expanding pattern towards creating the second era GM crops by exchanging esteem included yield qualities, fundamentally profiting customers and processors (OCTM, 2001; Azevedo and Welington 2003; ISO 2005 and 2006). The entry of sustenance DNA pieces over the intestinal divider is a characteristic and physiological marvel, chiefly when DNA is at high focuses in the nourishment. Given that domesticated animals expend significant measures of GM crop-inferred plant feed, open worry about the utilization of creature items containing transgenic DNA and protein have prompted examinations identified with their destiny inside the gastrointestinal tract of animals and the conceivable gathering of trans-qualities and their encoded proteins inside tissues, (Tylecote 2019, Kunling et.al 2019).
Endogenous and transgenic DNA pieces from low-duplicate qualities have been recognized in creature tissues, however in lesser sums than that distinguished on account of high-duplicate qualities. Section of plant DNA parts, endogenous or not, over the intestinal hindrance does not seem to have effectively affected animals (McClements,2019). Proteins communicated by GM crops have raised some worry as they might be associated with nourishment sensitivities. Surveying allergenicity to GM crops has additionally included physicochemical and biochemical measures relating protein strength to warmth, corrosive and stomach related chemicals. In addition, transgenic innovation has been helpful in creating hypoallergenic crops by meddling with the outflow of qualities encoding significant allergens. There is scientific confidence that GM crops do not represent greater risks than those already present in conventional agriculture and that any new risk posed by GM crops could be identified, managed and prevented (Zhiguang et al,2019, Wen-Ching et al.2019).
However, conventionally modified genes are numerous and their functions remain essentially unknown, whereas trans-genes are controlled by their nature, making them more reliable in terms of obtaining the desired outcome. Our aim is to detect the abnormalities in the GM rice varieties and the effect of it in the human being and also to modify the changes in a way so that the yield does not vary and also the effect on human being is not harmful. This work was designed to modify the crops without harming the body.
Materials and Methods
Materials
Every traditional variety has some nutritive or medicinal property. Today unfortunately, we have narrowed our choices to a handful of varieties and consume them polished devoid of fiber and minerals. We, instead, eat a range of processed foods made with the same grains and mistake variety for diversity. However, it is diversity that will give us a range of nutritional elements – apart from fulfilling our need for varied taste. Each of the different rice varieties have their particular taste profile, and often, most people develop their personal favorites. Two GM rice samples, FR0502519 and FR0502520, were kindly provided agriculture extension department of Centurion University. The samples were labelled as “Anti-pest Shanyou 63” and as “Anti-pest Jinyou 63”.
Sample material and DNA extraction
DNA was separated from ground material utilizing a changed CTAB nucleic corrosive extraction technique (ISO,2006). 1.5 mL CTAB extraction cradle (20 g/L cetyl-trimethyl-ammonium bromide, 1.4 mol/L NaCl, 0.1 mol/L Tris– HCl, 0.02 mol/L Na2EDTA, pH 8.0) and 10 μL Proteinase K arrangement (20 mg/mL) were added to 200 mg of the processed rice test. The examples were brooded at 60 °C under steady fomentation medium-term and afterward centrifuged for 10 min at 13,000×g. The supernatant was moved into another vial, 750 μL of chloroform was included, vivaciously shaken and after that centrifuged at 13,000×g for 5 min. The upper stage was moved into another vial, its volume was resolved and blended with two volumes of CTAB precipitation support (5 g/L cetyl-trimethyl-ammonium bromide, 40 mmol/L NaCl). After hatching for 60 min at room temperature without tumult, the examples were centrifuged for 15 min at 13,000×g, the supernatant was disposed of and the pellet was resuspended in 350 μL of a 350 mmol/L NaCl arrangement. Chloroform (350 μL) was included, the examples were blended on a Vortex and centrifuged for 10 min at 13,000×g. The upper stage was joined with 0.6 volumes of isopropanol for nucleic corrosive precipitation and after 20 min brooding at room temperature the examples were centrifuged 10 min at 13,000×g. The supernatant was disposed of, the pellet was washed with 500 μL ethanol arrangement (70% EtOH) and settled in 200 μL TE support (1 mmol/L Tris– HCl, 0.1 mmol/L Na2EDTA, pH 8.0).
The separated DNA was evaluated in a fluorometric examine utilizing the PicoGreen dsDNA restricting color (Invitrogen) as per the producer’s guidelines. Estimations were led in an ABI PRISM 7900 (Applied Biosystems) at 525 nm. A 100 bp sub-atomic size DNA marker with a centralization of 0.1 μg/μL DNA (Fermentas) was utilized for adjustment. For explicitness tests, DNA was removed from two financially accessible regular rice brands purchased in Germany (‘Wurzener Parboiled Reis’, Lot L 60531E, Wurzen, Germany and ‘Gut and Günstig Spitzen-Langkorn-Reis’, Lot 0722008, Hamburg, Germany), from GM rice line LLRICE62, GM soya line GTS40-3-2 and GM maize lines T25, Bt11, Bt10, Bt176, MON810, MON863, NK603, TC1507, GA21 and CBH351.
For affectability tests, a sequential weakening of DNA extricated from tests FR0502519 and FR0502520 was set up by stepwise fourfold weakening with 0.1× TE cushion. Another sequential weakening likewise used to test for the affectability was set up similarly with DNA extricated from test FR0502519 and a blend of genomic DNA from ordinary rice (‘Wurzener Parboiled Reis’, 10 μg/mL) and maize (10 μg/mL). The mass portion blends with various GMO substance were readied utilizing ground customary rice and GM rice test FR0502519. A blend of 9.5 g traditional rice and 0.5 g of rice test FR0502519 bringing about a 5% (w/w) test was utilized a beginning material. 2 g of this 5% (w/w) blend was added to 8 g of regular rice giving a 1% (w/w) test. A consequent 0.5% (w/w) blend was readied utilizing 5 g of the 1% (w/w) test and 5 g of customary rice, and a 0.1% (w/w) level was readied utilizing 8 g ordinary rice and 2 g of the 0.5% (w/w) test. For consequent quantitative continuous PCR investigations DNA was extricated from the 5, 0.5 and 0.1% mass part blends.
PCR and DNA sequencing
PCR was performed in a volume of 25 μL containing 2.5 μL 10× PCR support with 15 mmol/L MgCl2, 0.5 μmol/L of every preliminary, 0.625 U Taq polymerase (HotStar, Qiagen) and 1 μL of layout DNA relating to 25 ng DNA. For warm cycling, an underlying denaturation venture for 15 min at 95 °C was trailed by 45 cycles of 30 s at 94 °C, 30 s at 60 °C and 60 s at 72 °C with a last prolongation venture of 7 min at 72 °C. In traditional PCR and constant PCR focusing on the CaMV 35S advertiser and the nos eliminator groupings conventions portrayed somewhere else were utilized (Ehlers et.al, 1997).
For arrangement assurance of the transgenic develop DNA separated from tests FR0502519 and FR0502520 was utilized in PCR explores different avenues regarding groundworks RiceActin1f (5′-ccc tct cct ctt tct ttc tcc g-3′; individual correspondence N. Hess) in blend with groundwork NOS180R (5′-TTg TTT TCT ATC gCg TAT TAA ATg T-3′; individual correspondence R. Reiting) at response conditions as depicted. Extra PCR items were created with oligonucleotides NOS-1 (5′-gAA TCC TgT TgC Cgg TCT Tg-3′), NOS-3 (5′-TTA TCC TAg TTT gCg CgC TA-3′) and CryIA(b)F (5′-ACC ATC AAC AgC CgC TAC AAC gAC C-3′) utilizing response conditions depicted somewhere else (Ehlers et.al, 1997).
PCR items were decontaminated with a QIAquick PCR refinement unit (Qiagen) and legitimately sequenced with the BigDye Terminator V 1.1 cycle sequencing pack (Applied Biosystems) in an ABI PRISM 310 instrument (Applied Biosystems). Sequencing was finished by a groundwork strolling procedure to produce successions in covering areas of the diverse develop components. Nucleic corrosive succession information were first contrasted and the Sequence Navigator programming (Applied Biosystems) and afterward investigated via looks in the GenBank arrangement database utilizing the PC calculation BLAST 2.
Real-time PCR
Real Time PCR was performed in an ABI PRISM 7900 instrument (Applied Biosystems). All responses were kept running as copies in 96-well plates. The 25 μL response blends contained 12.5 μL all inclusive ace blend (Applied Biosystems), the showed convergences of groundworks and test (Table 1) and 5 μL of format DNA. The response conditions were as per the following: Initiation venture for 10 min at 95 °C pursued by 45 cycles of 20 s at 95 °C and 1 min at 60 °C. Preliminaries T51F and T51R1 and test T51p (Table 1) were structured with the Primer Express 2.0 programming (Applied Biosystems). Groundwork and test successions for location of taxon-explicit rice reference qualities were taken from distributed continuous PCR measures.
Table 1: Description of the different real-time PCR systems used in the study. The length of the generated PCR product is given in brackets
| Method | Name | Oligonucleotide sequence (5′–3′) | Final concentration in PCR (nmol/L) |
| Bt rice construct (83 bp)
|
T51F | gAC TgC Tgg AgT gAT TAT CgA CAg A | 400 |
| T51R | AgC TCg gTA CCT CgA CTT ATT CAg | 400 | |
| T51p | FAM-TCg AgT TCA TTC CAg TTA CTg CAA CAC TCg Ag-TAMRA | 200 | |
| gos9 rice reference gene (68 bp)
|
org1 | TTA gCC TCC CgC TgC AgA | 400 |
| org2 | AgA gTC CAC AAg TgC TCC Cg | 400 | |
| orgp | FAM-Cgg CAg TgT ggT Tgg TTT CTT Cgg-Dabcyl | 200 | |
| sps rice reference gene (81 bp) | SPSF | TTg CgC CTg AAC ggA TAT | 650 |
| SPSR | Cgg TTg ATC TTT TCg ggA Tg | 550 | |
| SPSP | FAM-gAC gCA Cgg ACg gCT Cgg A-Dabcyl | 200 |
Results and Discussion
Sequence analysis of GM rice samples
Two rice seed tests taken at neighborhood Chinese wholesalers by Greenpeace in the year 2005 were utilized for DNA arrangement examination. These examples, FR0502519 and FR0502520, were at first examined with GMO screening tests. The two examples demonstrated positive responses in a CryIA(c) immuno-based Bt-protein test and in a DNA-based test for the nopaline synthase (nos) translation eliminator arrangement, though just a single example (FR0502520) was certain for the CaMV 35S advertiser succession in a 35S DNA-test. Re-examination of these outcomes with a 35S advertiser explicit continuous PCR test demonstrated that just a powerless 35S response with a Ct-estimation of ∼35 is discernible with DNA from FR0502520 when contrasted with the Ct-estimation of ∼24 acquired in the nos-explicit constant PCR utilizing a similar measure of test DNA. We accept that hints of another GMO is available in test FR0502520, since the build in the suspected ‘GM Shanyou 63’ line ought not contain the CaMV 35S advertiser and the expulsion of the 35S advertiser driven hph marker quality by isolation has been accounted for the parental CMS restorer line ‘Minghui 63’.
A few distinctive groundwork blends focusing on the assumed transgenic build embedded in ‘GM Shanyou 63’ were utilized to create PCR items reasonable for direct DNA sequencing (Fig. 1). The nucleotide (nt) successions of these items were broke down with the BLAST likeness internet searcher of the National Center for Biotechnology Information (NCBI). The two locales chose for arrangements of the transgenic groupings with indistinguishable GenBank successions are appeared. 1 and 2). The 5′ succession piece of the enhanced part indicates total character to the rice Act1 intron contained in a plant change vector (pPLEX-5013) over a stretch of 404 nucleotides (Fig. 1). This grouping is trailed by a 38 nt long spacer with parts of a various cloning site until a potential ATG begin codon of the Bt poison quality encoding perusing outline is found. The following 347 nt significant lot indicates personality to an engineered CryIA(c) quality (increase number Y09787). The succession of the PCR items produced with DNA from FR0502519 and FR0502520 demonstrated no distinctions in this area (information not appeared), showing that either the two examples speak to a similar GM rice occasion, or that the two examples get from change occasions with a similar build. The distinguished transgenic develop apparently compares to plasmid pFHBT1 which was utilized for creation of the ‘GM Shanyou 63’ line. Especially a short amino-terminal peptide succession (P-N-I-N-E-C-I) erased in the half and half cryIA(c) quality of pFHBT1 is likewise not present in the reasoned protein arrangement of the distinguished perusing outline.
 |
Figure 1 |
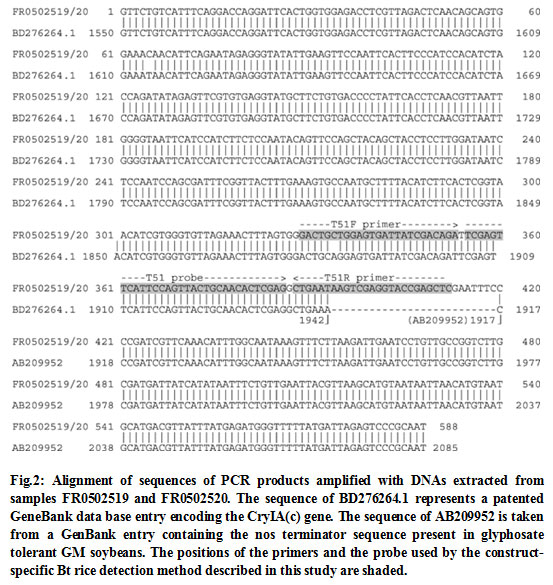 |
Figure 2 |
To additionally break down the transgenic groupings in the rice test materials under scrutiny, it was tried whether the nos eliminator is likewise situated behind the Bt cryIA(b) and cryIA(c) combination quality as portrayed for the pFHBT1 build. Utilizing DNA from FR0502519 and FR0502520 as layout, particular PCR items were enhanced with preliminaries spreading over the district of the 3′ part of the cryIA(c) quality and the nos eliminator. Sequencing results demonstrates that in this district no distinctions are available in the two explored rice tests (Fig. 2). The initial 393 nucleotides of the succession totally match to a GenBank database passage coding for an engineered cryIA(c) quality. Toward the finish of the cryIA(c) grouping homology a 26 nt long spacer with no similitude to realized successions is found, trailed by a 169 nt long arrangement indistinguishable to the nos eliminator got from Agrobacterium tumefaciens.
Crop-to-crop gene flow
All in all, developed rice is portrayed by high rates of self-fertilization and almost no cross-fertilization between nearby plants or fields (ordinarily under 1 percent). Tests in Italy demonstrated that dust intervened quality stream from a transgenic, herbicide-safe rice assortment to adjoining plants of a nontransgenic partner was 0.05 to 0.53 percent. Present day rice cultivars are regularly become close more established landraces (privately adjusted assortments that were trained and improved by customary ranchers) in Asia, and hybridization rates between these two gatherings additionally seem, by all accounts, to be extremely low . These discoveries are steady with the little separations that are suggested for detaching and keeping up the virtue of developed rice developed in seed nurseries. In the United States, for example, rice plants that are developed for ensured seed to be sold to ranchers must be segregated from other rice assortments by just 6 meters (m) or less.
Crop-to-weed gene flow
Another vital yet little-examined part of quality stream is the ingenuity of yield qualities following crop– weed hybridization. Similarly as half and half life is seen when ingrained, developed lines are crossed to deliver “mixture” rice, so may weedy rice profit by hybridizing with the harvest, if this outcomes in more prominent heterozygosity. I a work it has found that more prominent power in crop– weed cross breeds than in their weedy guardians, and frequencies of yield alleles in weedy rice were as high as 52 percent after just two years of contact with the harvest. In any case, in Arkansas, original cross breeds among developed and weedy rice blossomed so late that they had much lower wellness than their weedy guardians. Along these lines, the developmental significance of half breed energy in weedy rice populaces gives off an impression of being variable and ought to be examined all the more extensively.
Through the span of a few ages, crop qualities that are firmly pernicious to weedy rice, just as different qualities that are connected to malicious harvest qualities, are probably going to be cleansed from weedy populaces by normal choice and choice weights from ranchers. Then again, connected qualities that are related with more prominent survival and propagation are required to increment in recurrence following scenes of hybridization.
Spread of transgenic herbicide resistance
Transgenic herbicide opposition is a quality that could without much of a stretch be procured by weedy rice. Weed control in rice fields is progressively subject to herbicides in both created and creating nations, halfway in light of the fact that more established strategies for hand transplanting youthful rice plants into overflowed fields are being supplanted by direct seeding methods. This progress has brought about more awful issues with weeds, since weed seedlings can smother the development of rice seedlings. Rice handle that become intensely pervaded with weedy rice can end up unusable, in light of the fact that the weed is a successful copy of the harvest and its seemingly perpetual seed bank makes it exceptionally hard to destroy. Consequently, rice producers who can manage the cost of the expense of herbicides are anxious to embrace herbicide-safe rice assortments, despite the fact that the advantages of this methodology could be brief.
Effects of other fitness-related transgenes
As a rule, different transgenes are not expected to continue and spread in weedy or wild populaces. On the off chance that the transgenes encode qualities that don’t upgrade the plants’ survival or multiplication, the departure of these qualities is probably not going to result in natural issues, in light of the fact that the new qualities will be uncommon. In like manner, if the transgenes give a wellness cost, as diminished survival or fertility, people bearing these qualities will be more averse to pass them on to their descendants. Numerous transgenic characteristics identified with wholesome quality and grain arrangement are probably going to have nonpartisan or negative consequences for the wellness of weedy and wild relatives of the yield.As opposed to these precedents, the wellness of wild and weedy rice populaces may be improved by transgenes that give better vermin opposition, more noteworthy resistance of abiotic stresses, for example, dry spell and saltiness, and upgraded yields. Contingent upon nearby conditions, these transgenes may discharge weedy or wild populaces from natural weights that confine their neighborhood plenitude or breaking point their territory prerequisites.
A few interrelated inquiries emerge in regards to the natural impacts of wellness upgrading transgenes. To start with, given these qualities’ capability to spread and endure, their conceivable negative impacts on non target species ought to be considered. Would the transgene or its items lead to hurtful impacts on gainful bugs, natural life, or different species, and how might these impacts contrast and conceivable mischief coming about because of traditional farming practices.This lead us to the different practices which are causing hazards to the body. This MSA provide us the data of the genes affected in GM crop and also the data of the genes affected by the GM crops in human from which we can identify the genetic rectifications and the cause of it and through which we can stop the genetical modification in human which actually increasing day by day.
Conclusion
Taking everything into account, it merits stressing that quality stream is a characteristic procedure that happens unavoidably under normal conditions. Based on current natural and transformative learning, we trust that the dispersal of numerous sorts of transgenes from rice won’t be unsafe to the earth. Transgenes that don’t have critical common specific focal points should cause constrained ecological effect, assuming any. Then again, transgenes that lead to more prominent bounties of weedy or wild rice could present natural issues, as depicted previously. Before allowing the business arrival of new sorts of transgenic rice, policymakers should most likely gauge the net natural advantages and dangers of such transgenes against different elements, for example, the foreseen impacts of transgenic assortments on human wellbeing, nearby flourishing, and exchange. Natural evaluations that are deductively thorough and freely available are fundamental for the long haul accomplishment of this imperative innovation.
Funding
Centurion University of Technology and Management Odisha
References
Alexander Tw, Reuter T, Aulrich K, et al. (2007). A review of the detection and fate of novel plant molecules derived from biotechnology in livestock production. Anim Feed Sci Tech. 2007;133:31-62.
Andrew Tylecote (2019). Biotechnology as a new techno-economic paradigm that will help drive the world economy and mitigate climate change. Research Policy, Volume 48, Issue 4, Pages 858-868
Brookes G, Barfoot P (2006). GM crops: The first ten years -Global socioeconomic and environmental impacts. P.G. Economics.
Bucchini L, Goldman Lr (2002). StarLink corn: a risk analysis. Environ Health Perpect. 110:5-13.
Chakraborty S, Chakraborty N, Datta (2000) A. Increased nutritive value of transgenic potato by expressing a nonallergenic seed albumin gene from Amaranthus hypochondriacus. Proc Nat Acad Sci USA. 97(7):3724-3729.
David Julian McClements (2019), Food Biotechnology: Sculpting Genes with Genetic Engineering, Future Foods pp 261-286.
Fischer R, Emans N (2000). Molecular farming of pharmaceutical proteins. Transgenic Res. Volume;9:Page Number279-99.
Halpin C (2005). Genes stacking in transgenic plants-the challenge for 21st century plant biotechnology. Plant Biotechnology J. Volume;3: Pages 141-155.
Hodgson E (2001). Genetically modified plants and human health risks: Can additional research reduce uncertainties and increase public confidence? Toxicology Sci.;Volume 63:Pages153-156.
ILSI (2003). Best practices for the conduct of animal studies to evaluate crops genetically modified for input traits. International Life Sciences Institute, Washington, DC. 62 p..
ILSI (2004). Nutritional and safety assessments of foods and feeds nutritionally improved through biotechnology. Compr Rev Food Sci F. Volume 3:Pages: 36-104.
ISAAA (2006). Global status of commercialized biotech/GM crops: 2006. ISAAA Brief 35
ISO 21571: (2005) Foodstuffs. Methods of analysis for the detection of genetically modified organisms and derived products – nucleic acid extraction. International Standardization Organization, Geneva, Switzerland.
ISO 21569: (2005). Foodstuffs – methods of analysis for the detection of genetically modified organisms and derived products – qualitative nucleic acid based methods. International Standardization Organization, Geneva, Switzerland
ISO 21570: (2006). Foodstuffs – methods of analysis for the detection of genetically modified organisms and derived products – quantitative nucleic acid based methods. International Standardization Organization, Geneva, Switzerland
Jeong S-C, Pack Is, Cho E-Y, et al (2007). Molecular analysis and quantitative detection of a transgenic rice line expressing a bifunctional fusion TPSP. Food Control. Volume 18:Pages: 1434-1442.
João Lúcio Azevedo and Welington LuizAraujo (2003). Genetically modified crops: environmental and human health concerns. Volume 544, Issues 2–3, Pages 223-233.
Kari E. Dunfield and James J. Germida (2004). Impact of Genetically Modified Crops on Soil- and Plant-Associated Microbial Communities. Journal of Environmental Quality Abstract. Vol. 33 No. 3.
Kunling Chen,Yanpeng Wang, Rui Zhang, et.al (2019). CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture..Annual Review of Plant Biology. Vol. 70 Pages:667-697
Made D, Degner C, Grohmann L (2006). Detection of genetically modified rice: a construct-specific real-time PCR method based on DNA sequences from transgenic Bt rice. Eur Food Res Technol. Volume 224: Pages 271-278.
Margulis C (2006). The hazards of genetically engineered foods. Environ Health Persp.;Volume: 114 Issue(3):Pages:146-A147.
Munoz-Furlong A, Sampson Ha, Sicherer Sh (2004). Prevalence of self-reported seafood allergy in the U.S. J Allergy Clin Immunol.;Volume113(S):Pages: S100.
Newell-Mcgloughlin M (2006). A retrospective prospective perspective on agricultural biotechnology ten years on. J Comm Biotechnol. Volume 13:Pages 20-27.
Oliver C, Hankins J (2007). Future world leader in GM crops. The China Business Review. July-August. chinabusinessreview.com. p. 36-39.
Official Collection of Test Methods (2001) Screening for detection of genetically modified DNA sequences in foodstuffs by detection of DNA sequences which are frequently present in genetically modified organisms German Federal Foodstuffs Act – Food Analysis, Article 35, L 00.00-31.
Orlando Acosta1, Alejandro Chaparro (2008), genetically modified food crops and public health. Acta biol.colomb. Vol.13 no.3
Poulsen M, Kroghsbo S, Schrøder M,et al (2007). A 90-day safety study in Wistar rats fed genetically modified rice expressing snowdrop lectin Galanthus nivalis (GNA). Food Chem Toxicol.Volume;45:Pages 350-363.
Raney T, Pingali P (2007). Sowing a gene revolution. Sci Am. Volume 297 Issue (3):Pages:104-111.
Reece Walters (2004). Criminology and Genetically Modified Food. The British Journal of Criminology, Volume 44, Issue 2, Pages 151-167.
Schrøder M, Poulsen M, Wilcks A, et al (2007). A 90-day safety study of genetically modified rice expressing Cry1Ab protein (Bacillus thuringiensis toxin) in Wistar rats. Food Chem Toxicol. Volume 45:Pages 339-349.
Setamou M, Bernal Js, Legaspi Jc,et.al (2002). Parasitism and location of sugarcane borer (Lepidoptera: Pyralidae) by Cotesia flavipes (Hymenoptera: Braconidae) on transgenic and conventional sugarcane. Environ Entomol. Volume 31:Pages: 1219-1225.
Singh Ov, Ghai S, Paul D,et.al (2006) Genetically modified crops: success, safety assessment, and public concern. Appl Microbiol Biotechnol. Volume 71:Pages 598-607.
Taylor Sl, Goodman Re, Hefle Sl (2001). The development of safety assessment for genetically modified foods. Asia Pacific Biotech News. Volume10, Issue11,Pages:614-616.
Tolstrup K, Andersen S, Boelt B, et al (2003). Report from the Danish working group on the co-existence of genetically modified crops with conventional and organic crops. Ministry of Food, Agriculture and Fisheries-Danish Institute of Agricultural Sciences, Copenhagen.
Uzogara Sg (2000). The impact of genetic modification of human foods in the 21st century: A review. Biotechnol Adv. Volume18,Pages:179-206.
WAL JM. (2001). Biotechnology and allergic risk. Rev Fr Allergol Immunol Clin. Volume 41,Pages :36-41.
Wen-Ching Chen, Tai-Ying Chiou, Aileen L. Delgado (2019).The Control of Rice Blast Disease by the Novel Biofungicide Formulations. Volume 11 Page: 3449
Zhiguang Qiu Eleonora Egidi Hongwei Liu (2019). New frontiers in agriculture productivity: Optimised microbial inoculants and in situ microbiome engineering. Biotechnology Advances,Volume 37, Issue 6,


