1Institute of Veterinary Science and Animal Husbandry, SoA University, Odisha, India and ICAR-DFMD, Mukteswar,
Nainital, India and FAO Ref Centre for FMD in South Asia, Bhubaneswar, India.
2Former Vice Chancellor, Sardar Vallabhbhai Patel University of Argricultue &Technology, Meerut, India.
3Department of Studies in Food Technology, Davangere University, Shivagangotri, Davangere, India.
4Department of Biotechnology and Bioinformatics, School of Life Sciences, JSS Academy of
Higher Education & Research, Mysuru, India.
5Department of Sciences, Amrita School of Arts and Sciences, Amrita Vishwa Vidyapeetham,
Mysuru Campus, Mysuru, India.
6Department of Biotechnology, Davangere University, Shivagangotri, Davangere, India.
7ICAR-National Institute of Veterinary Epidemiology and Disease Informatics
(NIVEDI), Yelahanka, Bengaluru, India.
Corresponding author email: Sharanspin13@gmail.com
Article Publishing History
Received: 12/07/2022
Accepted After Revision: 21/09/2022
Vertebrate RNA viruses cause most of the infectious, contagious, transboundary diseases of mammalians in the world. Since 2000, H5N1 avian flu, H1N1/H1N2 Swine flu, SARS, MERS, CCHF, and Covid-19 have caused outbreaks. In addition, rabies, HIV, measles, viral hepatitis, respiratory viruses, dengue, in human beings, and FMD in cattle, PPR in goat, Bluetongue in sheep, infectious bronchitis and PRRS in pigs, are prevalent and endemic in different countries since many years. Whole virus inactivated and live attenuated vaccines including non-pathogenic mutants have been successful in control and eradication of many viral diseases of man and animals. The advancements in molecular virology, non-replicating designer virus vectors of adenovirus and Poxvirus origins, and replicating designer virus vectors derived from vesicular stomatitis virus are being used to deliver immunogenic genes of other viruses to confer protection.
RNA vaccine in the form of nucleoside modified mRNA has been successful in control of Covid-19. Single cycle replicon (SCR) virus construct with target transgene, and codon-pair bias deoptimized (CPD) virus have been promising vaccine platforms; both mutants are live attenuated and non-transmissive between host cells. Antigenic spectrum of CPD-virus is as wide as virus attenuated by serial passage in experimental hosts, and is very quick to develop. The technique of synthetic attenuated virus engineering has been faster in developing new age viral vaccines, and also faster to update to match antigenic diversity. This review describes applications of CPD, SAM and SCR technologies in developing vaccine candidates for RNA virus diseases.
Codon-Pair Deoptimization, New Age Vaccines, Self-Amplifying Mrna, Single Cycle Replicon,
Pattnaik B, Yadav M. P, Vidya G, Dharamashekara C, Shreevatsa B, Shivamallu C, Bhavana H. H, Kollur S. P, Srinivasa C, Patil S. S. New Age Vaccines: Technologies in Developing Vaccine Candidates for RNA Virus Diseases. Biosc.Biotech.Res.Comm. 2022;15(3).
Pattnaik B, Yadav M. P, Vidya G, Dharamashekara C, Shreevatsa B, Shivamallu C, Bhavana H. H, Kollur S. P, Srinivasa C, Patil S. S. New Age Vaccines: Technologies in Developing Vaccine Candidates for RNA Virus Diseases. Biosc.Biotech.Res.Comm. 2022;15(3). Available from: <a href=”https://bit.ly/3CJ2cpL“>https://bit.ly/3CJ2cpL</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Vaccines against various viral diseases have been proved to be saviour and guardian of one health domain since decades. Conventionally, two types of vaccines viz., live attenuated and inactivated vaccines have occupied the health sector with their own limitations as they are easy to produce comparatively. Cleverly viral agents have co-evolved in the vaccinated hosts and evaded the host immune mechanism. This has opened the newer areas of concern leading to revisit the vaccinology and produce more sophisticated vaccines using latest molecular biology techniques (Pollard and Bijker 2021). With this background, this review is thought of to impart knowledge of new age vaccines to the academicians and researchers.
Application of different platforms/ technologies like codon pair bias deoptimized (CPD), self-amplifying mRNA (SAM), single cycle replicon (SCR) vaccines, in developing viral vaccines has been described in detail by (Frederiksen et al. 2020). Still needs to be strengthened in bringing out the products using such technologies towards the use by stakeholders replication-deficient virus are genetically defective in replicating their genome, as in case of current adenovirus vectored SARS-CoV-2 spike gene vaccines, whereas in SCR-virus, the genome replicates using viral RNA replicase- transcriptase to increase copy number of the transgene(s)/ cloned gene(s) so that the target protein molecule is translated in more quantity in the host cells leading to higher immune response in the body, but there is no synthesis of new virion particles due to absence of critical viral gene(s) (Pollard and Bijker 2021).
The SCR virus technology is a quick and precise genomic process in attenuating pathogenic virus for use in vaccines, and en-capsidation (morphogenesis)- defective SCR- Flavivirus has been used as vaccine candidate (Widman et al. 2008; Terasaki et al. 2015). The SCR- virus mutant do not yield new virions, unlike the CPD attenuated virus. Incorporation of underrepresented (rare) codon pairs, but not frequency of CpG di-nucleotide, is the chemistry of attenuation by CPD (Groenke et al. 2020).
Of late, RNA vaccines in the form of self-amplifying mRNA (SAM), alternatively self-amplifying RNA (saRNA), built on alphavirus replicon, has found wide application including for control of Covid-19 pandemic Frederiksen et al. 2020; Bloom et al. 2021). Some of the examples of pathogenic RNA viruses modified as vaccine candidates by nucleotide sequence alteration/ deletion technologies, viz., CPD, SAM and SCR are presented in the present review for the benefit of academicians and researchers in virology and vaccinology. Vaccine candidates developed by either of these three new technologies are classified as live attenuated and non-infectious (Pollard and Bijker 2021).
Genome composition in vertebrate RNA viruses: The relative occurrence of dinucleotides (two adjacent nucleotides in a polynucleotide chain) is genomic signature of a species Elango et al. 2009). The di-nucleotide TpA is rare (under-represented) in most of the organisms to circumvent nonsense mutations, whereas frequency of occurrence of CpG vary between eukaryotes. Use of CpG in (-) single stranded RNA viruses may not be influenced by the host tropism. In contrast to single stranded RNA viruses and reverse transcribing ones, there is high occurrence of CpG di-nucleotide in dsRNA viruses (Karlin and Mrazek 1997; Cheng et al. 2013).
CPD attenuated virus as vaccine candidate: For codon optimization, synonymous replacement of rare codons is made to match with availability of codon specific aminoacyl-tRNA in host cells that enhances protein translation. Inclusion of rare codons attenuated poliovirus (Burns et al. 2006; Mueller et al. 2006), and this attenuation process was named as synthetic attenuated virus engineering (Coleman et al. 2008). Codon pair bias in virus is host species dependent, and virulent virus can be attenuated by synonymous replacement of efficient codon pairs with inefficient/ rare codon pairs in the required viral gene (protein) that is called as Codon-pair bias deoptimization with no alteration in amino acid sequence and composition (Coleman et al. 2008; Broadbent et al. 2016; Kunec and Osterrieder 2016).
Codon-pair (bias) deoptimization (CPD) has been effectively used to attenuate virulent viruses (Coleman et al. 2008; Mueller et al. 2010; Broadbent et al. 2016; Groenke et al. 2020). CPD attenuates viruses as genes re-encoded with rare codon pairs translate less protein due to limitation in availability of required codon specific aminoacyl-tRNAs in host cells; protein formation is controlled by the quantity of mRNA copies and their translatability that is linked to availability of tRNA isotypes (Broadbent et al. 2016; Groenke et al. 2020). The CPD virus is replication competent, antigenically identical to the parent strain, and induce specific immune response akin to the virulent strain (Coleman et al. 2008; Mueller et al. 2010; Broadbent et al. 2016).
Self-amplifying mRNA/RNA (SAM/saRNA) as vaccine candidate: During the last about two decades, vaccine development platform using synthetic RNA has been adapted with success; mRNA vaccines for preclusion of infectious diseases have been a substitute for conventional vaccines, with the additional advantage of being cold-chain independent (Maruggi et al. 2019; Bloom et al. 2021). The RNA/mRNA vaccines are commonly formulated on RNA genome of single stranded positive sense RNA viruses (Maruggi et al. 2019; Bloom et al. 2021). RNA vaccine platform using synthetic alphavirus replicon that has 5′ cap, NSP1–4, 26S sub-genomic promoter, and 3′ poly A tail, but no structural protein genes, known as self-amplifying mRNA (SAM)/ self-amplifying RNA (saRNA) technology, has been promising (Zhou et al. 1994; Geall et al. 2012; VanderVeen et al. 2012; Geall et al. 2012; Luis et al. 2015; Samsa et al. 2018; Ballesteros-Briones et al. 2020).
RNA vaccines on alphavirus derived SAM stimulates innate immunity through pattern recognition receptors (PRRs), and also elicits strong and specific humoral and cellular immune responses (Yoneyama et al. 2010; Ulmer et al. 2012; Atasheva et al. 2012; Maruggi et al. 2019; Ballesteros-Briones et al. 2020). SAM vaccines using influenza virus HA, human cytomegalovirus glycoprotein gB, HIV envelope glycoprotein, respiratory syncytial virus F antigen, malaria- plasmodium protein PMIF, rabies virus G protein have been developed and evaluated (Geall et al. 2012; Brazzoli et al. 2015; Brito et al. 2015 Bogers et al. 2015; Baeza Garcia et al. 2018; Stokes et al. 2020).
Single-cycle replicon (SCR) virus as vaccine candidate: Application of SCR technology in developing vaccine candidate for RNA virus diseases and also for ds-DNA viruses like ASFV and HSV is described (Freitas et al. 2019; Ramsey et al. 2020).
(i) Single-strand segmented negative sense RNA virus: Rift Valley Fever virus (RVFV): Rift valley fever (RVF) is a zoonotic disease primarily of domestic ruminants and humans with an incubation period of 2-6 days (WHO) that was first documented in 1931 in the Great Rift Valley of Kenya. The RVFV is an insect-borne enveloped virus (Fig:1) that is maintained between ruminants and mosquitoes. Humans get infected by the RVFV either by mosquito bite or contact with materials contaminated with the virus.
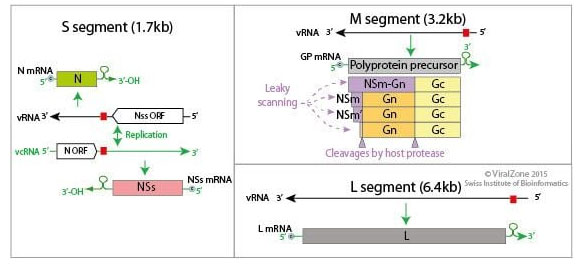
The virus to the genus Phlebovirus, under family Phenuiviridae and order Bunyavirales, and has 3 segments of negative sense RNA genome (11.5 kb), viz., L, M, and S (Walter and Barr 2011; Nuss et al. 2014) (Fig: 2). The genus phlebovirus has more than 60 species (ICTV, 2021). As the RNA genome segments are of negative sense, the complete virion particles carry molecules of L and N proteins that are required for the initial replication/ transcription of viral RNA (Nuss et al. 2014).
Figure 1: Structure of RVF virus. vRNP- viral ribo-nucleoprotein, three in number; N- nucleoprotein;
L- viral polymerase; Gn and Gc- virus surface glycoproteins.
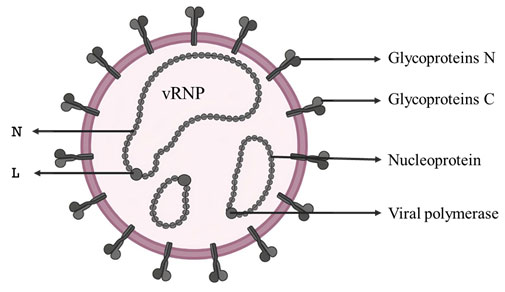
Figure 2: Genome organisation of RVF virus (Source: Alchetron: Phlebovirus)
Immunization with RVF-VLP carrying N gene protected mice (Pichlmair et al. 2010). VLPs are devoid of viral genome, whereas SCR virions are genomic mutant. SCR-RVFV elicited protective immunity without side effects in hosts (Terasaki et al. 2015). This SCR-RVFV mutant had L and S RNAs, and devoid of M RNA; absence of M RNA segment (no glycoproteins Gn and Gc) prevented formation of new virus particles. The L and N gene/protein in the SCR-RVFV effected single-cycle replication of the mutant RVFV genomes, followed by translation and accumulation of L and N proteins in the infected cells.
An SCR-RVFV having NSm, Gn and Gc gene sequences protected mice and sheep (Kortekaas et al. 2012). Another SCR- RVFV carrying nucleotide sequences of L- RNA, N gene, and Gn protected mice and provided sterile immunity in sheep after single dose; Gn glycoprotein elicited high level of neutralizing antibodies in vaccinates (Oreshkova et al. 2013). SCR-RVFV prepared from a mutant strain MP-12 and having L-RNA, M-RNA with mutations F826N and N827A in Gc glycoprotein to eliminate membrane fusion activity of the viral envelope required during morphogenesis of new virions and prevent multi cycle infection, and N gene (S-RNA) protected suckling mice after intracranial challenge with MP-12 RVFV (Terasaki et al. 2015). A new, promising vaccine for RVF is significantly SCR-MP-12 (Caplen et al. 1985; Murakami et al. 2016).
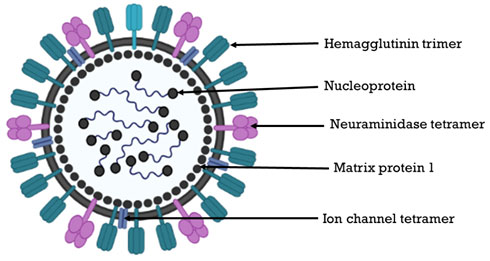
Influenza A virus: Influenza A virus (IAV) is the only species of genus Alpha influenza virus in the family Orthomyxoviridae. A, B, C, and D are the four primary influenza viral types. A big strain of influenza A virus infects wild aquatic birds and can spread to other terrestrial species, including humans. IAVs, like all avian influenza viruses (AIVs), are enveloped, pleomorphic, and have eight distinct negative sense RNA genomic regions extending from 890 to 2341 nucleotides (Webster et al. 1992; Klenk et al. 2008). RNA polymerase complex PB2 (cap-binding), PB1+F2 (polymerase) and PA (endonuclease), (HA) hemagglutinin (segment 4), NP (segment 5), neuramidase (NA) (segment 6), M1 and M2 (segment 7) and NS1+NEP (segment 8) are the viral proteins
(Fig: 3). Nucleoprotein (NP) coats each RNA genome segment, forming a ribonucleoprotein (RNP) complex with RdRp and many copies of nucleoprotein (NP) (Lo et al. 2018). The hemagglutinin and neuramidase glycoproteins on the virus surface are antigenically diverse, and divide the IAVs into 18 H and 11 N antigenic subtypes, respectively (Joshi et al. 2021). It includes two new subtypes each of hemagglutinin and neuramidase (H17N10, H18N11) lately identified in bats. The Influenza A serotypes that have been approved in humans are, H1N1, H1N2 (endemic in humans, pigs and birds), H2N2, H3N2, H5N1 (bird flu/ avian flu), H6N1, H7N2, H7N3, H7N7, H7N9, H9N2, and H10N7 (Joshi et al. 2021).
Figure 3: Structure of Influenza virus (Source: The virology down under blog)
In the upper respiratory tract, the virus adheres to sialic acid (SA) linked to galactose molecule by alpha- 2,6 linkage (SA-alpha2,6 Gal), and to SA linked to galactose by alpha- 2,3 linkages (SA-alpha2,3 Gal) in the lower respiratory tract (Shinya et al.2006). Human adenovirus vector platform has been used for influenza A vaccines (Van Kampen et al. 2005; Weaver et al. 2009; Weaver 2014; He et al. 2015). Adenovirus serotype 6 was engineered to a SCR- virus, having the E1 gene but deleted E3 gene cassette to prevent formation of new virion particles, to express HA gene of influenza A/PR/8/34 virus (Crosby et al. 2017).
The expression cassette consisting of HA gene with CMV promoter- enhancer and SV40 poly(A), and was cloned between Ad6 fibre and E4 genes. In Syrian hamsters and cotton rats, the SCR-Ad6-HA construct generated high level of anti- hemagglutinin antibodies (hemagglutination inhibition), equal to 50% protective dose in humans. Cotton rats vaccinated intranasally developed high level of anti-HA antibodies within 21 days, with low viral load in lungs one day post challenge with live A/PR/8/34 virus at 48 days post vaccination. These observations suggest that SCR-adenovirus based vaccines could be suitable for a number of viral pathogens (Crosby et al. 2017). An SCR-Ad6 virus has been used to develop vaccine for Influenza A virus (Crosby et al. 2017; Anguiano-Zarate et al. 2018).
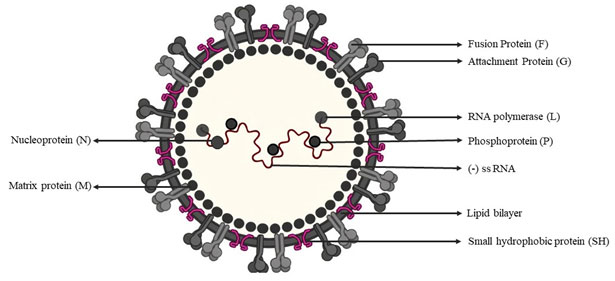
Single strand negative sense RNA virus: Respiratory syncytial virus (RSV): Acute infection of lower respiratory tract by RSV is a global human health problem with no licensed vaccine (Mazur et al. 2018). RSV belongs to genus Orthopneumovirus in family Pneumoviridae, order Mononegavirales (ICTV). The virion (120-200nm in size) contains one molecule of negative-sense, single-stranded RNA genome of about 15.2 kb with genome organisation 5’-NS1-NS2-N-P-M-F-M2(1,2)-SH-G-L-3’ (Munir et al. 2018; Cifuentes-Muñoz and Dutch 2019). The RNA genome has 10 ORFs coding 11 proteins viz., 5’→3’, NS1, NS2, N, M, P, G, F, SH, M2-1, M2-2 and L (Lee et al. 2012; Rincheval et al. 2017).
There are two overlapping ORFs in the M2 mRNA that yield two distinct matrix proteins, M2-1 and M2-2 (Borchers et al. 2013). The virus isenveloped with surface glycoproteins F, G and SH, the F and G are essential for virus attachment and fusion (Fig:4). The glycoprotein F mediates cell fusion to form pathognomonic multi-nucleated syncytium, and antigenically and phylogenetically classify RSVs in to types A and B; the former is more virulent than RSV-B (Jha et al. 2016). The other structural proteins are Nucleoprotein (N), Phosphoprotein (P), L (viral RNA polymerase) and matrix (M). NS1 and NS2 suppress type I interferon production and signalling (Munir et al. 2018).
Figure: 4. Structure and composition of RSV (Source: biggiesboxers.com)
RSV replicates in the cytoplasm of host cells, and there is formation of spherical cytoplasmic inclusion bodies. RSV infection causing respiratory disease in younger children was identified in 1956, and since then only two RSV-antibody preparations, RSV-IVIG and palivizumab, have been licensed for prevention (Mazur et al.2018). Difficulty in having an appropriate vaccine has been due to incomplete understanding of the immunology of RSV, and therefore, different vaccine technologies, viz., nanoparticle-based, live attenuated/ chimeric, virus subunit, vector-based platforms have been applied, and different vaccines for neonate, children and adult were developed (Mazur et al. 2018).
Formalin inactivated whole virus vaccine had side effects of respiratory disease (Kim et al. 1969; Mazur et al. 2018). Full-length F- glycoprotein- nanoparticle vaccine (Novavax 2015-18) was partially protective in adults (Mazur et al. 2018). So also, the GSK-RSV-F subunit vaccine had the problem of instability of the pre-F antigen. Further, an F glycoprotein subunit vaccine with a TLR4 agonist as adjuvant, was not efficient in eliminating respiratory sickness (Falloon et al. 2017). DPX-RSV vaccine developed using DepoVax technology and SH protein of RSV was promising (Karkada et al. 2010; Schepens et al. 2015).
A live attenuated RSV vaccine candidate was developed by CPD (Nouen et al. 2014). Vaccine formulation containing F protein attached to empty bacterial particles was not successful (Van Braeckel-Budimir et al. 2013; Mazur et al. 2018). MVA (modified vaccinia virus Ankara; replication deficient virus) vectored RSV vaccine (MVA-BN-RSV) expressing F, G, N and M2-1 antigens, and replication deficient chimpanzee Adenovirus 155 carrying viral antigens F, N, and M2-1 have been developed and under evaluation. Human Ad26-RSV-PreF vaccine candidate is also under evaluation. VXA-RSV-F, an adenovirus 5 based oral tablet, is expected to circumvent immunosenescence in adult people. Live attenuated RSV with deletion of either M2-2 or NS2 gene elicited strong neutralizing antibodies in children (Karron et al. 2013; Karron et al. 2015).
The rBCG-N-RSV chimeric vaccine expressing nucleoprotein of the virus elicited specific antibody and Th1 response essential for protection of lungs (Rey-Jurado et al. 2017). Recently, a (deletion) mutant SCR-RSV strain lacking matrix (M) protein gene was developed (Schmidt et al. 2018). The M protein is essential for new virion assembly/ morphogenesis. Infection with RSV M-negative mutant do not produce M protein, and therefore no new virion is generated during replication. The SCR-RSV lacking M gene, induced robust serum antibody and memory T cell response in mice. This SCR-RSV is a promising live attenuated vaccine candidate, as it provided protection in mice against live virus challenge and reduced virus replication in lung (Schmidt et al. 2019).
EBOLA virus (EBOV): The genus Ebolavirus belongs to family Filoviridae in the order Mononegavirales (ICTV). Virion appears filamentous under electron microscope (Fig:5), and contains one molecule of single-stranded negative-sense RNA. There are 7 genes, flanked by UTRs at both the ends. The virus genome is organised as: 3′-UTR-NP-VP35-VP40-GP-VP30-VP24-L-5′-UTR (Feldmann and Geisbert 2011; Schmidt et al. 2019).
Figure 5: Electron micrograph of Ebola virus (Source: CDC/Cynthia Goldsmith – Public Health Image Library, #10816).
Ebola virus disease (EVD) is a highly fatal viral haemorrhagic fever that was identified in 1976 in Sudan and Congo, near the Ebola River (WHO 2014). The virus spreads through infected body fluids including blood, and incubation period varies from 2-21 days. Four of the viruses in the genus Ebolavirus, viz., Bundibugyo virus, Sudan virus, Taï Forest virus, and Ebola virus are associated with Ebola virus disease (Hoenen et al. 2012). Genome back bone of both negative-sense single stranded RNA virus (vesicular stomatitis virus; VSV) and double-stranded DNA virus (ChAd3) were used to clone glycoprotein of EBOV; leading to replication competent rVSV-EBOV and replication defective ChAd3-EBOV vaccine candidates (Holbrook 2018; Samai et al. 2018; Suder et al. 2018).
The rVSV-EBOV is a live chimera in which G protein of VSV is replaced with EBOV-G protein. As the ChAd3-EBOV vaccine candidate was replication deficient, there was limited production of the target EBOV-G protein. Both the vaccine candidates were efficacious, but the rVSV-EBOV vaccine candidate caused severe side effects in recipients during clinical trial. MVA-EBOV-G recombinant vaccine candidate could be a booster for ChAd3-EBOV vaccine (Stanley et al. 2014; Holbrook 2018). A SCR-Adenovirus 6 platform carrying EBOV-G protein elicited strong humoral antibody response in mice, hamsters and rhesus macaques, and protected them from rVSV-EBOV challenge (Anguiano-Zarate et al. 2018).
Single- strand positive sense RNA virus: Flavivirus: The genus Flavivirus in the family Flaviviridae have single-stranded (+) sense RNA genome of about 10-11 kb with 5’ cap, but no 3’ Poly A tail (ICTV). Flavivirus is about 40 nm in diameter with icosahedral capsid, enveloped with surface projections of 5-10 nm, and replicate in host cell cytoplasm (Chambers et al. 1990). The RNA genome is infectious and monocistronic, that is a single polyprotein is translated from the RNA genome that is proteolytically processed by viral and host proteases to form individual viral proteins (Widman et al. 2008).
The members of flavivirus genus are Yellow fever, Dengue, Zika, Japanese Encephalitis, and West Nile Virus encephalitis etc that are transmitted by mosquito/ tick. Several subunit vaccines were also developed and used. Purified NS1 of YFV reduced viraemia in rabbits (Schlesinger et al.1986). Viral protein subunit vaccines were satisfactory for Dengue, JE and WNE (Widman et al. 2008). Several virus-vectored vaccines for Flavivirus diseases were developed and evaluated. Immunization with recombinant Vaccinia and Canarypox virus carrying structural and non-structural proteins of Flavivirus elicited protective immune response in laboratory animals and rhesus monkey (Bray et al. 1989; Yasuda et al. 1990; Konishi et al. 1998; Raengsakulrach et al. 1999; Iglesias et al. 2006).
Lentivirus (positive sense RNA virus) vector carrying trE gene of WNV was protective against live virus challenge after vaccination with 50 recombinant virus particles (Iglesias et al. 2006). Measles virus (negative sense RNA virus) vector carrying E gene of WNV elicited strong neutralizing antibody response and spared mice from live virus challenge (Despres et al. 2005). Replication-defective Adenovirus vectors have been used to express flavivirus antigens, and virus vector carrying NS-1 of TBEV protected > 50% of mice (Jacobs et al. 1994). Adenovirus carrying E gene of Dengue virus-2 elicited neutralizing antibodies in mice (Jaiswal et al. 2003).
Adenovirus vector carrying DIII of E gene of both Dengue virus-2 and 4 elicited virus neutralizing antibodies in laboratory animals (Khanam et al. 2007). A formulation carrying two Adenovirus recombinant vectors carrying M and E genes of four serotypes of Dengue virus elicited virus neutralizing antibody response against all the 4 serotypes (Raja et al. 2007; Holman et al. 2007). So, the platform was successful for WNV (Schepp-Berglind et al. 2007). DNA based vaccines for Flavivirus diseases were developed and evaluated (Widman et al. 2008). Plasmid carrying M and E genes of Dengue virus- 2 elicited virus neutralizing antibody response in mice (Konishi et al. 2000; Widman et al. 2008).
DNA construct carrying either M and E genes or NS-1 of JE virus protected 70-90% of mice (Lin et al. 1998). Simultaneous administration of DNA vaccine carrying M and E genes of WNV, and formalin inactivated WNV vaccine had synergistic effect on neutralizing antibody response (Ishikawa et al. 2007). Multivalent recombinant plasmid/ DNA carrying M and E genes of the four Dengue virus serotypes elicited neutralizing antibody response against all (Konishi et al. 2006; Raviprakash et al. 2006). A DNA vaccine for WNV carrying M and E genes was effective in mouse and horse and turned out to be the first DNA vaccine licensed for use in animals (Davis et al. 2001; Widman et al. 2008).
Dengue virus attenuated by deletion of a part in the 3’UTR elicited neutralizing antibodies in human and non-human primates (Durbin et al. 2001). Flaviviruses have the potential to exchange structural genes amongst members of the same genus (Widman et al. 2008). The C-prM-E regions of Dengue virus 1 and 3 was cloned in Dengue virus 4 backbone, and the live chimeras were more attenuated than the parent virus (Bray and Lai 1991). ChimeriVax vaccine platform carrying M and E genes of the virus cloned in YFV-17D backbone has been promising (Widman et al. 2008).
The M and E genes of JEV were cloned in YFV-17D backbone (Chambers et al. 1999). This chimera, ChimeriVax-JE, was immunogenic and safe in humans (Monath et al. 2002). ChimeriVax-Dengue was constructed for all Dengue virus serotypes, and ChimeriVax-Dengue-2 elicited strong neutralizing antibody response (Guirakhoo et al. 2006). Viral E protein of WNV was mutated to reduce neurovirulence in mice, and mutated ChimeriVax-WNV (ChimeriVax-WN02) was constructed that protected immunized monkeys from viraemia (Arroyo et al. 2004). SCR- TBE virus was designed by deletion in the Capsid (C) gene of the virus; this attenuated mutant (Kofler et al. 2004) as well as synthetic viral RNA with same deletion elicited defensive immunity in adult mice (Aberle et al. 2005).
This is the first report of RNA vaccine in the form of SCR- Flavivirus. It was shown that YF and WN virus genome with large deletion in the capsid gene can be packaged in host cells expressing the capsid protein from another related virus replicon (Mason et al. 2006). RepliVAX is a Capsid gene deletion mutant virus that replicated and produced progeny virions in cells expressing capsid protein from a replicon of the Venezuelan equine encephalitis (VEE) virus (genus Alphavirus; Togaviridae). However, the mutant genome may replicate once (SCR) in other cells without supplementing the missing capsid protein. RepliVAX-WNV vaccine was protective in mice and hamster (Widman et al. 2008). A RepliVAX- JEV chimera was constructed to prevent JE (Ishikawa et al. 2008).
Alphavirus: Members of Genus Alphavirus, family Togaviridae, carry positive sense RNA genome of 11-12 kb with 5’Cap and 3’ Poly A tail (ICTV). The virus is enveloped with icosahedral capsid core, and about 70nm in diameter. There are 2 ORFs in the viral RNA genome; the ORF 1 is towards 5’ end of the genome that codes for four non-structural proteins, NSPs1-4, whereas the ORF2 is located towards the 3’ end of the genome and codes for structural proteins, viz., capsid (C), E3, E2, 6K and E1. The E3 and E2 originate from the precursor protein P62.
There are three subgroups in the genus Alphavirus; Semliki Forest virus subgroup, eastern equine encephalitis virus subgroup (EEEV and VEEV) and the Sindbis virus subgroup (Levinson et al. 1990). VEEV is a mosquito-borne viral pathogen that affect all equine species, and also is an emerging zoonotic agent. There are 6 different subtypes, I to VI, of VEEV. The available vaccines include, TC-83 live attenuated and C-84 formalin inactivated vaccines. These vaccines have side effects. Viral RNA genome vaccine platform stimulates innate/ non-specific immune system and antigen expressed by RNA vaccine elicits strong and specific immune response (Ulmer et al. 2012; Atasheva et al.2012). The alphavirus replicon technology (that gave rise to SAM technology) has been utilised in the development of vaccination for animals and man (VanderVeen et al. 2012).
RNA vaccine platform using SAM was developed that used a 9 kb RNA in lipid nano particles (Geall et al. 2012). This SAM construct was a synthetic RNA representing Alphavirus genes coding for RNA replicase/ transcriptase but lacked structural protein genes. This construct included 5′ cap, NSP1–4, 26S sub-genomic promoter, the target gene/protein/antigen (transgene), and 3′ poly A tail. SAM vaccine with VEEV strain TC-83 genome devoid of capsid gene was developed (Samsa et al. 2018). This synthetic SAM at 100μg dose elicited virus neutralizing antibodies, similar to those elicited by TC-83 live attenuated VEE vaccine, and provided thorough protection in mice. This VEE-SAM vaccine is live-attenuated, and undergoes RNA amplification without production of new virion (Samsa et al. 2018).
Coronavirus: Spike gene of Infectious Bronchitis Virus (IBV) was found to be a candidate for new IBV vaccine, as Thymidine Kinase deficient recombinant vaccinia virus carrying Spike (S) gene of IBV elicited virus neutralizing antibodies in mice (Tomley et al. 1987). This has led to Spike gene-based vaccines for SARs-CoV-2, TGEV, and CCoV (Torres et al. 1995; Qiao et al. 2005; Yuan et al. 2015; Frederiksen et al. 2020; Lu et al. 2021). Recombinant NDV, LaSota strain, having complete S gene of IBV, cloned between phosphoprotein (P) and matrix (M) genes of NDV, protected chicks against IBV (Abozeid et al. 2019; Jbeli and Jelassi 2021).
Virus-independent entry/ delivery of nucleic acids in to host cells has advantage over virus vectored delivery system (Geall et al. 2012). Replication-deficient virus, whose genome does not replicate upon infection of susceptible host cells, has been used to formulate different viral vaccines. The gene insert/ transcription unit/transgene is transcribed and translated, and as the recombinant viral DNA does not replicate the antigen/ protein yield could be limited. In contrast, single cycle replicon (SCR) virus, the genome of which replicates with no progeny virion formation, has been used in vaccine design. In both the cases, the mutant virus is classified as live attenuated, and non-infectious as the mutant(s) cannot spread in the body of the host, and whole virus can be rescued only in cells supplementing the missing viral component.
The SCR- mutant based vaccine has the capability and advantage, over replication-defective virus vaccine, of multiplying the viral genome and multiple genome copies produced (by SCR mutant) would lead to higher number of transcribed mRNAs leading to translation of high level of viral proteins, and thereby higher immune response in the host (Widman et al. 2008). Therefore, SCR- vaccines would be superior to replication-deficient vaccines, and can be designed for both RNA and DNA viruses. Further, SCR- vaccine is an easier and efficient platform for positive-sense RNA viruses. The principle of codon-pair bias deoptimization (CPD) by synthetic attenuated virus engineering (SAVE), has revolutionized virus attenuation. Quick development of attenuated virus strains by CPD-SAVE has facilitated faster development of live virus vaccines.
Pathogenic viruses attenuated by CPD can be used as vaccine candidates as such, or can be further modified (recombination) by insertion of an alien gene/ transcription unit (vaccine target) in the inter-genic space available (e.g. between phosphoprotein and matrix genes, and Nucleoprotein and phosphoprotein genes etc) in negative-sense single stranded RNA viruses, viz., morbillivirus, orthoavulavirus and vesiculovirus etc (Brandler et al. 2007; Kasama et al. 2011; Mok et al. 2012; Jbeli and Jelassi 2021). Live-attenuated Schwarz strain of Measles Virus developed by passage in primary human kidney and amnion cells, and then in CEF cells can be used to express heterologous antigens and is being used to develop measles virus- based Covid-19 vaccine (Combredet et al. 2003; Boisgerault et al. 2013; Frederiksen et al. 2020).
Recombinant Measles virus-based SARS-CoV-2 vaccine candidate carrying full length Spike gene of SARS-CoV-2 were found highly efficacious (Horner et al. 2020; Lu et al. 2021). Avirulent Newcastle disease virus (NDV; Orthoavulavirus, Paramyxoviridae) carrying Spike gene of infectious bronchitis virus (IBV; coronavirus) was protective in chicks (Abozeid et al. 2019). The CPD technique is also being applied to develop live attenuated vaccine candidates for SARS-CoV-2 (Frederiksen et al. 2020).
The SAM/saRNA vaccine technology using SCR- Alphavirus replicon/ vector has been a success in different virus models including SARS-CoV-2 on VEEV replicon (Zhou et al. 1994; Geall et al. 2012; Vander Veen et al. 2012; Sandbrink and Shattock 2020; Bloom et al. 2021). The transgene (target gene/ transcription unit) is amplified by the Alphavirus RdRp complex for higher antigen/ protein production without formation of progeny virion; the SAM vaccine is also classified as live-attenuated. These are usually delivered encapsulated in lipid nano particles (Geall et al. 2012; Bloom et al. 2021).
CONCLUSION
The findings of the present review have shown that the development of RNA virus vaccine candidates on RNA virus backbone, e.g., (1) attenuated negative-sense single stranded RNA virus (e.g., measles virus, NDV, VSV etc) carrying a transgene, (2) CPD virus designed by SAVE, and (3) SAM/saRNA molecule designed on alphavirus replicon, is gaining wide application as these vaccine technologies/ platforms are easier to adapt, and such new age viral vaccines can be development in less time, compared to whole virus vaccines, both inactivated and live attenuated, and DNA virus (Adenovirus and Poxvirus) vectored vaccines.
Conflict of Interests: Authors declare no conflict of interests to disclose.
REFERENCES
Aberle, J. H., Aberle, S. W., Allison, S. L., et al. (1999). A DNA immunization model study with constructs expressing the tick-borne encephalitis virus envelope protein E in different physical forms. Journal of Immunology. 163(12):6756–6761.
Abozeid H H, Anandan Paldurai, Varghese B P et al. (2019). Development of a recombinant Newcastle disease virus-vectored vaccine for infectious bronchitis virus variant strains circulating in Egypt. Vet Res 50:12. https://doi.org/10.1186/s13567-019-0631-5.
Anguiano-Zarate S S, Matchett W E, Nehete P N, et al. (2018). A Replicating Single-Cycle Adenovirus Vaccine Against Ebola Virus. The Journal of Infectious Diseases; 218:1883–89.
Atasheva, S., Akhrymuk, M., Frolova, E.I., et al. (2012). New PARP gene with an anti-alphavirus function. J. Virol. 86, 8147–8160.
Baeza Garcia, A., Siu, E., Sun, T., et al.. (2018). Neutralization of the Plasmodium-encoded MIF ortholog confers protective immunity against malaria infection. Nature Communications. 9, 2714.
Ballesteros-Briones M C, Noelia Silva-Pilipich, Guillermo Herrador-Cañete, et al. (2020). A new generation of vaccines based on alphavirus self-amplifying RNA. Current Opinion in Virology 44:145-153,https://doi.org/10.1016/j.coviro.2020.08.003.
Bloom K, van den Berg F and Arbuthnot P (2021). Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2021 Apr;28(3-4):117-129. doi: 10.1038/s41434-020-00204-y. Epub 2020 Oct 22. PMID: 33093657; PMCID: PMC7580817.
Bogers WM, Oostermeijer H, Mooij P, et al. (2015). Potent immune responses in rhesus macaques induced by nonviral delivery of a self-amplifying RNA vaccine expressing HIV type 1 envelope with a cationic nanoemulsion. Journal Infected Diseases. 2015, 211:947-955.
Brandler S, Marianne Lucas-Hourani, Arnaud Moris et al. (2007). Pediatric Measles Vaccine Expressing a Dengue Antigen Induces Durable Serotype-specific Neutralizing Antibodies to Dengue Virus. PLOS Neglected Tropical Diseases. 1(3): e96. doi:10.1371/journal.pntd.0000096.
Brazzoli M, Magini D, Bonci A, et al. (2015). Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. Journal Virology. 2016, 90:332-344.
Broadbent A J, Celia P. Santos, Amanda Anafu, et al. (2016). Evaluation of the attenuation, immunogenicity, and efficacy of a live virus vaccine generated by codon-pair bias de-optimization of the 2009 pandemic H1N1 influenza virus, in ferrets. Vaccine 34(4): 563-570. https://doi.org/10.1016/j.vaccine.2015.11.054
Caplen, H., Peters, C.J and Bishop, D.H (1985). Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. Journal of General Virology. 66 (Pt 10), 2271–2277.
Chambers T J, Hahn C S, Galler R, et al. (1990). Flavivirus genome organization, expression, and replication. Annual Review Microbiology. 1990;44:649-88. doi: 10.1146/annurev.mi.44.100190.003245. PMID: 2174669.
Cheng, X., Virk, N., Chen, W., et al. (2013). CpG usage in RNA viruses: data and hypotheses. PloS one, 8(9), e74109. https://doi.org/10.1371/journal.pone.0074109
Coleman J R, Dimitris Papamichail, Steven Skiena et al. (2008). Virus Attenuation by Genome-Scale Changes in Codon Pair Bias. Science 27 Jun 2008:Vol. 320, Issue 5884, pp. 1784-1787.DOI: 10.1126/science.1155761
Crosby C M, Matchett W E, Anguiano-Zarate S S et al. (2017). Replicating Single-Cycle Adenovirus Vectors Generate Amplified Influenza Vaccine Responses. Journal of Virology. 91: e00720-16. https://doi.org/10.1128/ JVI.00720-16.
Feldmann H and W Geisbert T W (2011). Ebola haemorrhagic fever. Lancet 2011; 377: 849–62. DOI:10.1016/S0140- 6736(10)60667-8.
Frederiksen LSF, Zhang Y, Foged C et al. (2020). The Long Road Toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies. front immunology. 2020 Jul 21;11:1817. doi: 10.3389/fimmu.2020.01817. PMID: 32793245
Freitas F B, Margarida Simões, Gonçalo Frouco, et al. (2019). Towards the Generation of an ASFV-pA104R DISC Mutant and a Complementary Cell Line—A Potential Methodology for the Production of a Vaccine Candidate. Vaccines 2019, 7, 68; doi:10.3390/vaccines7030068.
Geall A J, Verma A, Otten G R, et al. (2012). Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci U S A 2012, 109:14604-14609.
Groenke N, Trimpert J, Merz S et al. (2020). Mechanism of Virus Attenuation by Codon Pair Deoptimization. Cell Reports.2020 Apr 28;31(4):107586. doi: 10.1016/j.celrep.2020.107586. PMID: 32348767.
He B, Zheng B-J, Wang Q, et al. (2015). Adenovirus-based vaccines against avian-origin H5N1 influenza viruses. Microbes and Infection 17:135–141. https://doi.org/10.1016/j.micinf.2014.11.003.
Holman, D. H., Wang, D., Raviprakash, K., et al. (2007). Two complex, adenovirus-based vaccines that together induce immune responses to all four dengue virus serotypes. Clin. Vaccine Immunology. 14 (2):182–189.
Horner C (2020). A highly immunogenic and effective measles virus-based Th1-biased COVID-19 vaccine. PNAS 117 (51): 32657–32666. doi:10.1073/pnas.2014468117.
Ishikawa T, Widman DG, Bourne N, et al. (2008). Construction and evaluation of a chimeric pseudoinfectious virus vaccine to prevent Japanese encephalitis. Vaccine. 2008 May 23;26(22):2772-81. doi: 10.1016/j.vaccine.2008.03.010.
Jacobs, S. C., Stephenson, J. R., and Wilkinson, G. W. (1994). Protection elicited by a replication-defective adenovirus vector expressing the tick-borne encephalitis virus non-structural glycoprotein NS1. Journal of General Virology. 75(Pt 9):2399–2402.
Jbeli R and Jelassi A (2021). Current vaccine technology with an emphasis on recombinant measles virus as a new perspective for vaccination against SARS-CoV-2. Euro-Mediterr J Environ Integr. 2021;6(2):61. doi: 10.1007/s41207-021-00263-6. Epub 2021 Jul 4. PMID: 34250222; PMCID: PMC8254859.
Joshi, S., Parkar, J., Ansari, A., et al. (2021). Role of favipiravir in the treatment of COVID-19. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases. 102, 501–508. https://doi.org/10.1016/j.ijid.2020.10.069
Karlin, S., and Mrázek, J. (1997). Compositional differences within and between eukaryotic genomes. Proceedings of the National Academy of Sciences of the United States of America, 94(19), 10227–10232. https://doi.org/10.1073/pnas.94.19.10227.
Kasama Y, Satoh M, Saito M et al. (2011). Evaluation of a Recombinant Measles Virus as the Expression Vector of Hepatitis C Virus Envelope Proteins. World Journal of Vaccines, 1, 98-103. doi:10.4236/wjv.2011.13010.
Klenk H. (2008). Avian Influenza: Molecular Mechanisms of Pathogenesis and Host Range. Animal Viruses: Molecular Biology. Academic Press. (2008): 978-1-904455-22-6.
Kortekaas, J., Antonis, A.F., Kant, J., et al. (2012). Efficacy of three candidate Rift Valley fever vaccines in sheep. Vaccine 30 (23), 3423–3429.
Kunec D and Osterrieder N (2016). Codon Pair Bias Is a Direct Consequence of Dinucleotide Bias. Cell Reports 14(1): 55- 67, https://doi.org/10.1016/j.celrep.2015.12.011.
Lu M (2021). A safe and highly efficacious measles virus-based vaccine expressing SARS-CoV-2 stabilized prefusion spike. PNAS March 23, 2021 118 (12) e2026153118; https://doi.org/10.1073/ pnas.2026153118
Maruggi G, Zhang C, Li J, Ulmer JB et al. (2019). mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Molecular Therapy. 2019 Apr 10;27(4):757-772. doi: 10.1016/j.ymthe.2019.01.020. Epub 2019 Feb 7. PMID: 30803823; PMCID: PMC6453507.
Mason, P. W., Shustov, A. V., and Frolov, I. (2006). Production and characterization of vaccines based on flaviviruses defective in replication. Virology 351(2):432–443.
Mazur N I (2018). The respiratory syncytial virus vaccine landscape: lessons from the graveyard and promising candidates. The Lancet. Infectious diseases, 18(10), e295–e311. https://doi.org/10.1016/S1473-3099(18)30292-5
Mok H, Xing Cheng, Qi Xu et al. (2012). Evaluation of measles vaccine virus as a vector to deliver respiratory syncytial virus fusion protein or Epstein-Barr virus glycoprotein gp350, The Open Virology Journal, 2012, 6, 12-22.
Mueller S, Coleman J, Papamichail D et al. (2010). Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol 28, 723–726. https://doi.org/10.1038/nbt.1636.
Nuss, J.E., Kehn-Hall, K., Benedict, A., et al. (2014). Multi-faceted proteomic characterization of host protein complement of Rift Valley fever virus virions and identification of specific heat shock proteins, including HSP90, as important viral host factors. PLOS ONE 9 (5), e93483.
Oreshkova, N., van Keulen, L., Kant, J., et al. (2013). A single vaccination with an improved non-spreading Rift Valley fever virus vaccine provides sterile immunity in lambs. PLOS ONE 8 (10), e77461.
Pichlmair, A., Habjan, M., Unger, H., et al. (2010). Virus-like particles expressing the nucleocapsid gene as an efficient vaccine against Rift Valley fever virus. Vector Borne Zoonotic Diseases. 10 (7), 701–703.
Pollard A.J. and Bijker E.M. (2021). A guide to vaccinology: from basic principles to new developments. Nature Reviews: Immunology. 21 (2), 83-100. https://doi.org/10.1038/ s41577-020-00479-7
Qiao J, Xia XZ, Yang ST et al. (2005). Construction of recombinant canine adenovirus type 2 expressing Canine coronavirus spike glycoprotein and its immunogenicity. Wei Sheng Wu Xue Bao. 2005 Aug;45(4):588-92. Chinese. PMID: 16245877.
Raja, N. U., Holman, D. H., Wang, D., et al. (2007). Induction of bivalent immune responses by expression of dengue virus type 1 and type 2 antigens from a single complex adenoviral vector. American Journal of Tropical Medicine. 76(4):743–751.
Samsa M M, Dupuy LC, Beard CW, et al. (2018). Self-Amplifying RNA Vaccines for Venezuelan Equine Encephalitis Virus Induce Robust Protective Immunogenicity in Mice. Molecular Therapy 27(4). https://doi.org/10.1016/j.ymthe.2018.12.013.
Sandbrink J B and Shattock R J (2020). RNA Vaccines: A Suitable Platform for Tackling Emerging Pandemics? Front Immunology. 2020 Dec 22;11:608460. doi: 10.3389/fimmu.2020.608460. PMID: 33414790; PMCID: PMC7783390.
Schepp-Berglind, J., Luo, M., Wang, D., et al. (2007). Complex adenovirus-mediated expression of West Nile virus C, PreM, E, and NS1 proteins induces both humoral and cellular immune responses. Clin. Vaccine Immunology. 14(9):1117–1126.
Schlesinger, J. J., Brandriss, M. W., Cropp, C. B., et al. (1986). Protection against yellow fever in monkeys by immunization with yellow fever virus nonstructural protein NS1. Journal of Virology. 60(3):1153–1155.
Schmidt M E, Oomens AGP, and Varga SM(2019). Vaccination with a Single-Cycle Respiratory Syncytial Virus Is Immunogenic and Protective in Mice. Journal of Immunology. 2019; 202:3234-3245. doi: 10.4049/jimmunol.1900050.
Schmidt, M. E., Knudson C. J., Hartwig S. M. et al. (2018). Memory CD8 T cells mediate severe immunopathology following respiratory syncytial virus infection. PLoS Pathology. 14: e1006810.
Stokes A, Pion J, Binazon O, et al. (2020). Nonclinical safety assessment of repeated administration and biodistribution of a novel rabies self-amplifying mRNA vaccine in rats. Regulatory Toxicology and Pharmacology. 113:104648.
Terasaki K, Tercero B R and Makino S (2015). Single-cycle replicable Rift Valley fever virus mutants as safe vaccine candidates. Virus Research, 216: 55-65. https://doi.org/10.1016/j.virusres.2015.05.012.
Tomley F.M., Mockett A.P., Boursnell M.E., et al. (1987). Expression of the infectious bronchitis virus spike protein by recombinant vaccinia virus and induction of neutralizing antibodies in vaccinated mice, Journal of General Virology. 68:2291–2298. doi: 10.1099/0022-1317-68-9-2291. PMID: 2821170.
Ulmer, J.B., Mason, P.W., Geall, A., et al. (2012). RNA-based vaccines. Vaccine 30, 4414–4418.
Van Kampen K R, Shi Z, Gao P, et al. (2005). Safety and immunogenicity of adenovirusvectored nasal and epicutaneous influenza vaccines in humans. Vaccine 23:1029 –1036. https://doi.org/10.1016/j.vaccine.2004.07.043.
Vander Veen R L, Harris D L and Kamrud K I (2012). Alphavirus replicon vaccines. Animal Health Research Reviews. Jun;13(1):1-9. doi: 10.1017/S1466252312000011.
Walter C T and Barr J N (2011). Recent advances in the molecular and cellular biology of bunyaviruses. Journal of General Virology. 92 (Pt 11), 2467–2484.
Weaver E A (2014). Vaccines within vaccines: the use of adenovirus types 4 and 7 as influenza vaccine vectors. Human Vaccines & Immunotherapeutics.10: 544 –556. https://doi.org/10.4161/hv.27238.
Weaver E A, Nehete P N, Buchl S S et al. (2009). Comparison of replication-competent, first generation, and helper-dependent adenoviral vaccines. PLoS One 4:e5059. https:// doi.org/10.1371/journal.pone.0005059.
Webster R G, Bean W J, Gorman O T, et al. (1992). Evolution and ecology of influenza A viruses. Microbiological Reviews. 1992 Mar;56(1):152-79. doi: 10.1128/mr.56.1.152-179.1992. PMID: 1579108; PMCID: PMC372859.
Widman D G, Frolov I and Mason P W (2008). Third‐Generation Flavivirus Vaccines Based on Single‐Cycle, Encapsidation‐Defective Viruses, Advances in Virus Research, Academic Press, Volume 72, Chapter 2, 2008, Pages 77-126,ISSN 0065-3527,ISBN 9780123743220, https://doi.org/10.1016/S0065-3527(08)00402-8.
Yoneyama M and Fujita T (2010). Recognition of viral nucleic acids in innate immunity. Review in Medical Virology. 2010 Jan;20(1):4-22. doi: 10.1002/rmv.633. PMID: 20041442.
Zhou X, Berglund P, Rhodes G, et al. (1994). Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine 12 (16): 1510-1514. https://doi.org/10.1016/0264-410X(94)90074-4.


