1Department of Microbiology, Kamani Science & Prataprai Arts College, Saurashtra University, Amreli India
2Dean of Dairy Science College, Kamdhenu University, Amreli India
Corresponding author email: shradhdhasmit@gmail.com
Article Publishing History
Received: 15/04/2020
Accepted After Revision: 26/05/2020
The present study shows the successful isolation and identification of lactic acid bacteria from mother’s milk. A total of three lactic acid bacteria designated as M2P1, S1 and M2L2 were isolated on MRS medium. Morphological analysis revealed that M2P1 produces small, creamy white, entire, convex and opaque colonies, S1 produces small, off white, entire, raised and opaque colonies, while M2L2 produces medium, creamy white, round shaped, raised and opaque colonies. Microscopic observation reveled that all the isolates are Gram-positive. To confirm the molecular identity of isolates, 16S rRNA sequencing was performed. The isolates M2P1, S1 and M2L2 were identified as Lactobacillus fermentum, Lactobacillus oris and Lactobacillus fermentum, respectively. Further characterization of all three isolates to check their potential as probiotics is under progress.
Isolation; Lactic acid bacteria; Mother’s milk; Screening
Gondaliya S, Ramani V. Isolation, Screening and Identification of Lactic Acid Bacteria from Human Milk. Biosc.Biotech.Res.Comm. 2020;13(2).
Gondaliya S, Ramani V. Isolation, Screening and Identification of Lactic Acid Bacteria from Human Milk. Biosc.Biotech.Res.Comm. 2020;13(2). Available from: https://bit.ly/2KfMvtL
Copyright © Gondaliya and Ramani This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Milk is a prime source of nutrition for young mammals. It is considered as a complete food with all the nutrients. Colostrum is early-lactation milk which carries the mother’s antibodies to the baby and reduces the risk of many diseases in the baby. Fermentation of milk is the first technique developed by humans for food preservation and it has played many important roles in nutrition. Lactic acid bacteria (LAB) are non-taxonomic group of Gram-positive, non-spore forming, fastidious facultative anaerobes having low mol% G+C content (Zhu. et al., 2009; Taguchi et al., 2008; Mercenier et al., 2000). The genera Lactobacillus, Lactococcus, Leuconostoc, Pediococcus and Streptococcus are considered as food associated LAB (Makarova et al., 2006). They require certain specific nutrients such as carbohydrates, amino acids, peptides, vitamins etc for their growth,
(Dipak, & Sheela, 2015, Huidrom & Sharma, 2018, Adams & Gutiérrez, 2019 and Talashi & Sharma, 2019).
LAB have ability to survive when they passed through the gastrointestinal tract and under acidic conditions. Apart from that they are known as ‘generally regarded as safe (GRAS)’ lactic acid producers (Bilkova et al, 2008). Above properties make them suitable candidate for their use in various fermentation, food and pharmaceutical processes. Especially, in food industries, LAB is used to enhance the flavor and texture of the food in addition to preserve the fermented food. LAB includes various Lactobacillus species which are naturally indigenous microflora of the fermented milk products. In recent time interest of using various Lactobacillus species as probiotics (according to FAO report probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit on the host) is increased because they are resistant to antibiotics. In fact, probiotics have been used for as long as people started the use of fermented foods (Pangallo et al., 2008, Shin et al., 2019).
Probiotic bacteria show positive affect in the individual by enhancing the properties of the indigenous microflora and maintaining microintestinal balance. It also competes with disease causing bacteria for nutrients absorption and villi attachment sites. Use of probiotics have reduced symptoms of lactose intolerance, improved immune function, cholesterol lowering function, antimutagenic activity and treatment of diarrhea (Shu et al. 2017, Mahmoudi et al., 2018 and Lockyer and Stanner, 2019).
However, in 20th century, Elie Metchnikoff proposed that ingested bacteria could have a positive influence on the normal microbial flora of the intestinal tract. He suggested that Lactobacillus species are important for human health and longevity, and thus, he promoted the use of yogurt and other fermented foods as healthy diet (Metchnikoff , 2004). LAB of human origin is more suitable to be used as a source of probiotics because they are adapted to the normal conditions prevailing in the gastrointestinal tract and therefore they are more competitive than others. In addition to that some Lactobacillus species have shown inhibitory action against common human pathogens. Lactobacillus species are also a producer of bacteriocin which has therapeutic importance as well as acting as a good bio-preservative of food products (Mobarez et al., 2008). Looking towards these many benefits there is urgent need to develop a potential LAB culture having ability to act as a source of probiotics.By considering the above values in mind, the present study was designed for the isolation and screening of potential lactic acid bacteria from mother’s milk sample. After the isolation, a representative lactic acid bacterial culture was identified through 16S rRNA sequencing.
MATERIALS AND METHODS
Mother’s milk is a rich source of health beneficial microflora. So, in the present research mother milk was selected to isolate various strains of Lactic acid bacteria. Before isolation the milk sample was stored at low refrigerated temperature i.e. -4°C to retain the normal milk flora and avoid contamination. Primary screening of lactic acid bacteria from mother’s milk was performed by the enrichment of milk flora in de Man Rogosa Sharpe broth. For the enrichment of milk flora 1 mL of the milk sample was added into 9 mL of de Man Rogosa Sharpe (MRS) broth, and it was incubated at 37°C for 48h. For the secondary screening of lactic acid bacteria the enriched MRS broth was serially diluted, and plated on different MRS agar plates followed by the incubation of all plates at 37°C until visible colonies of bacteria were observed. Selected bacterial colonies which grew on MRS plates were streaked on fresh MRS agar plates to obtain pure cultures. The pure cultures were maintained in 15% skimmed milk at -4°C. To retain the culture in active state, they were regularly transferred to the freshly prepared MRS medium and then stored as mentioned above. The purity of isolated bacterial cultures was primarily done through morphological and microscopic observations.
The bacterial cultures were identified through 16S rRNA sequencing according to the protocol shown by (Sambrook, 2001; Ikram et al., 2016) with slight modifications. In brief, 1.0 mL of 12h grown bacterial culture in MRS broth was centrifuged at 10,000 x g for 2 min and then, the cell pellet was washed with the sterile deionized water. Later, the cell pellet was suspended in 0.2 ml of 10 mM Tris-HCl, PH 8.0, 10 mM EDTA, pH 8.0, 20 mM glucose, 50 mM Sodium chloride mixture containing lysozyme and incubated at 37°C for 30 min. Following to that 25 μL of proteinase K (20 mg/ml) and 100 μL of 10 % SDS were added to it and the mixture was incubated for 1 h at 60°C. After that 300 μl of 3M sodium acetate (pH5.2) was added to the mixture and kept on ice for 5 min to allow the precipitation of nucleic acid.
The cell debris was removed by centrifugation at 10,000×g for 15 min at 4°C, and then the supernatant was collected in separate sterile micro-centrifuge tube. In the supernatant an equal volumes of phenol-chloroform-isoamylalcohol (25:24:1 v/v) was added with gentle shaking. Later on the preparation was centrifuged at 15000×g for 10 min to separate out two phases. DNA containing upper aqueous layer was carefully transferred in sterile micro-centrifuge tube. In next step, DNA was precipitated by adding two volumes of ice-cold ethanol with gentle shaking, and then it was incubated at 4°C for 1 h. The mixture was again centrifuged at 15000×g at 4°C for 10 min. The pellet was then rinsed with 70% (v/v) ice-cold ethanol and dried it in air. Then the dried mass was dissolved in 50μL of deionized water. Following to that 10μL of 10 mg/mL RNase was added into the preparation and it was incubated at 37°C for 15 min. Finally, the sample was kept at 4°C for PCR reaction and at -15°C for long term usage.
16S rRNA gene sequencing using PCR
Single-pass sequencing was performed on each template using below 16s rRNA universal primers. The fluorescent-labeled fragments were purified from the unincorporated terminators with an ethanol precipitation protocol. The samples were resuspended in distilled water and subjected to electrophoresis in an ABI 3730xl sequencer (Applied Biosystems). The 16s rRNA sequence was blast using NCBI blast similarity search tool. Thephylogeny analysis of query sequence with the closely related sequence of blast resultswas performed followed by multiple sequence alignment. The program MUSCLE 3.7 was used for multiple alignments of sequences (Edgar, 2004). The resulting aligned sequences were cured using the program Gblocks 0.91b.This Gblocks eliminates poorly aligned positions and divergent regions (removes alignmentnoise) (Talavera and Castresana, 2007). Finally, the program PhyML 3.0 aLRT was used for phylogeny analysis and HKY85 as Substitution model. PhyML was shown to be at least as accurate as other existing phylogeny programs usingsimulated data, while being one order of magnitude faster. PhyML was shown to be atleast as accurate as other existing phylogeny programs using simulated data, while beingone order of magnitude faster. The program Tree Dyn 198.3 was used for tree rendering (Dereeper, 2008; Reis et al., 2016).
RESULTS AND DISCUSSION
After extensive screening a total of three different lactic acid bacteria were isolated on MRS agar plates. They were designated as M2P1, S1 and M2L2. On MRS agar plate isolate M2P1 produced small, creamy white, entire, convex and opaque colonies, S1 produced small, off white, entire, raised and opaque colonies, while M2L2 produced medium, creamy white, round shaped, raised and opaque colonies. Detailed colony characteristics of all three isolates are shown in Table 1. During their studies Baradaran et al. (2012) successfully selected three lactic acid bacteria designated as K1, K3 and K5 out of total 12 different isolates which were isolated from the leaves of Malaysian herb Kesum. In their study all three isolates were observed to produce colonies with round, convex and creamy white appearances which were similar to the isolates of our study. In another study, Chowdhury et al., (2012) isolated four different lactic acid bacteria from buffalo yogurt.
Table 1. Colony characteristics of lactic acid bacteria
| Colony character | M2P1 | S1 | M2L2 |
| Size | Small | Small | Medium |
| Shape | Round | Round | Round |
| Margin | Entire | Entire | Entire |
| Texture | Smooth | Smooth | Smooth |
| Elevation | Convex | Raised | Raised |
| Consistency | Moist | Moist | Moist |
| Pigmentation | Creamy white | Off white | Creamy white |
| Opacity | Opaque | Opaque | Opaque |
Microscopic observation of lactic acid bacteria
All three isolates namely, M2P1, S1 and M2L2 were studied for Gram’s reaction followed by microscopic observation. During microscopic observation M2P1, S1 and M2L2 observed as Gram-positive, rod shaped non-spore forming bacteria which indicated that they have characteristics of Lactic acid bacteria. Baradaran et al. (2012) shown that lactic acid bacteria designated as K1, K3 and K5 isolated from the leaves of Kesum were Gram-positive and catalase negative upon microscopic and biochemical characterization, respectively.
Identification of Lactic acid bacteria
Ben Amor et al. (2007) suggested that physicochemical and biochemical analysis based identification of lactic acid bacteria is often unreliable because many different bacterial species exhibit similar morphological and nutritional requirements. So, in our study, after microscopic observation all three isolates were identified through 16S rRNA gene sequencing. The 16s rDNA sequences were aligned through BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) (Tilhaun et al., 2018).
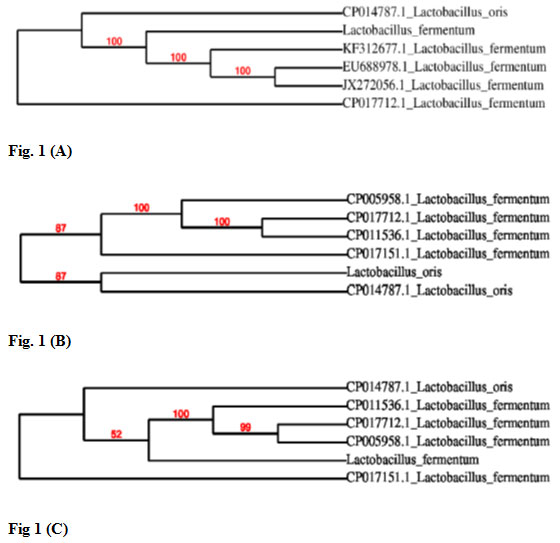
Later, sequences of all three isolates were compared with the published sequences of 16S rDNA gene of different lactic acid bacteria strains available at NCBI database. Based on 16S rRNA gene sequencing results bacteria designated as M2P1, S1 and M2L2 were identified as Lactobacillus fermentum, Lactobacillus oris and Lactobacillus fermentum, respectively. Results of phylogenetic analysis of all three isolates are shown in Fig. 1. On the basis of 16S rRNA sequencing results lactic acid bacteria K1, K3 and K5 were identified as Lactococcus lactis, Pediococcus pentosaceus and Lactobacillus curvatus, respectively (Baradaran et al., 2012).
Figure 1: Phylogenetic analysis of isolated lactic acid bacteria. Figure 1 (A) Based on phylogenetic analysis an isolate M2P1 was identified as Lactobacillus fermentum. Figure 1 (B) Based on phylogenetic analysis an isolate S1 was identified as Lactobacillus oris. Figure 1 (C) Based on phylogenetic analysis an isolate M2L2 was identified as Lactobacillus fermentum.
Several parameters have to be kept in mind while using the probiotics. These include type of prebiotics, concentrations, presence of other components, probiotic microorganism to be used, incubation time and fate of metabolism of prebiotics (Sharma et al., 2017; Singla and Chakkaravarthi, 2017).
CONCLUSION
The present research work highlights successful isolation and identification of lactic acid bacteria from mother’s milk. In the present study a total of three different bacteria namely, M2P1, S1 and M2L2 were isolated on MRS agar medium. All isolates were identified as Gram-positive bacteria through microscopic observations. Finally, on the basis of 16S rRNA sequencing results isolates M2P1, S1 and M2L2 were identified as Lactobacillus fermentum, Lactobacillus oris and Lactobacillus fermentum, respectively. Further characterization to check the potential of isolated lactic acid bacteria as probiotics is under progress.
REFERENCES
Adams CA, and Gutiérrez, B. (2019). Human Milk Oligosacharides: The First Prebiotics. Acta Scientific Nutritional Health, 3(12), 172–175. doi:10.31080/asnh.2019.03.0553
Baradaran A, Foo H. L, Sieo C. C and Rahim R. A. ( 2012) Isolation, identification and characterization of lactic acid bacteria from Polygonum minus. Romanian Biotechnol. Let. Vol. 17 No.3
Ben Amor K, Vaughan, E. and deVos W. (2007) Advanced molecular tools for the identification of lactic acid bacteria. J. of Nutrition Vol. 137 No.3: Pages 741
Bilkova A, Kinova Sepova H, Bilka F, Bukovsky M, Balazova A, Bezakova L. (2008) Acta Facultatis pharmaceuticae universitatis comeianae Vol. 55: Pages 64-72
Chowdhury A, Hossain M, Mostazir N. J, Fakruddin M, Billah M, and Ahmed M. M. (2012) Screening of Lactobacillus spp. from buffalo yoghurt for probiotic and antibacterial Activity. http://dx.doi.org/10.4172/2155-9597.1000156.
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM and Gascuel O. Phylogeny.fr (2008) robust phylogenetic analysis for the non-specialist. Nucleic Acids Res Vol. 1: Pages 36
Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. Vol. 32 No 5: Pages 1792-1797
Huidrom, S, and Sharma, N. (2018). Isolation and Screening of Novel Isolates of Bifido bacteria from Human Milk as Potential Probiotic with Antidiarrheal Activity. Annals of Applied Microbiology & Biotechnology Journal, 2(1), 1–8. doi:10.36876/aamb.1007
Ikram Medjaoui , Bouabdellah Rahmani , Malika Talhi , Fatima Zohra Mahammi , Fatima Zohra Moghtit , Nadhira Mehtar and Semir Bechir Suheil Gaouar (2016) Isolation and Characterization of Lactic Acid Bacteria from Human Milk and Newborn Feces . Journal of Pure and Applied Microbiology Vol. 10 No 4: Pages 2613-2620
Lockyer, S., & Stanner, S. (2019). Prebiotics – an added benefit of some fibre types. Nutrition Bulletin, 44(1), 74–91. doi:10.1111/nbu.12366
Mahmoudi, I., Telmoudi, A., & Hassouna, M. (2018). Beneficial Effects of Probiotic Lactobacillus plantarum Isolated from Cow, Goat and Sheep Raw Milks. Acta Scientific Microbiology, 1(2), 17–20. doi:10.31080/asmi.2018.01.0013
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E. (2006) Comparative genomics of the lactic acid bacteria. Proceeding National Academy of Sciences Vol. 103 No 42: Pages 15611
Mercenier, A., Muller-Alouf, H., and Grangette, C. (2000) Lactic acid bacteria as live vaccines. Curr. Issues Mol. Bio Vol. 2: Pages 17-26
Metchnikoff II. The prolongation of life: Optimistic studies. Springer Published Comp. New York (2004).
Mithun, S., Dipak, V., & Sheela, S. (2015). Isolation and Identification of Lactobacilli from raw milk samples obtained from Aarey Milk Colony. International Journal of Scientific and Research Publications, 5(1), 1–5.
Mobarez AM, Doust RH, Sattari M, Mantheghi N Antimicrobial (2008) effects of Bacteriocin like substance produced by L. acidophilus from traditional yoghurt on P. aeruginosa and S. aureus. J Biol Sci vol. 8: Pages 221-224
Reis N.A., Saraiva M.A.F., Duarte E.A.A., Carvalho E.A. de, Vieira B.B. and Evangelista-Barreto N.S. (2016) Probiotic properties of lactic acid bacteria isolated from human milk Vol. 121 : Pages 811-820
Pangallo D, Drahovska H, Harichova J, Karelova E, Chovancova K, Feriano p, Turna J, Timko J (2008) Antonie Van Leeuwenheok Vol. 94: Pages 555-562
Sambrook, J. and Russell, D. (2001) Molecular cloning: a laboratory manual: CSHL press
Sharma, P., Singh, A., & Mathur, N. (2017). Review Synbiotics, Combination of Probiotics And Prebiotics, IOSR Journal of Biotechnology and Biochemistry, 03(04), 19–24. doi:10.9790/264x-03041924
Shin, D., Chang, S. Y., Bogere, P., Won, K. H., Choi, J. Y., Choi, Y. J., … Heo, J. (2019). Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLoS ONE, 14(8), 1–23. doi:10.1371/journal.pone.0220843
Shu, G., He, Y., Chen, L., Song, Y., Meng, J., & Chen, H. (2017). Microencapsulation of Lactobacillus Acidophilus by xanthan-chitosan and its stability in yoghurt. Polymers, 9(12), 1–11. doi:10.3390/polym9120733
Singla, V., & Chakkaravarthi, S. (2017). Applications of prebiotics in food industry: A review. Food Sci Technol Int, 23(8), 649–667. doi:10.1177/1082013217721769
Taguchi, S., Yamada, M., Matsumoto, K., Tajima, K., Satoh, Y., Munekata, M. A (2008) microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme. Proceeding National Academy of Sciences Vol. 105 No 45: Pages 17323
Talashi, S, & Sharma, N. (2019). Isolation of Lactobacillus plantarum from Human Breast Milk with Probiotic and Medical attributes. Acta Scientific Microbiology, 2(6), 163–171. doi:10.31080/ASMI.2019.02.0258
Talavera, G. and Castresana, J. (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology Vol. 56: Pages 564-577
Tilahun, B., Tesfaye, A., Muleta, D., Bahiru, A., Terefework, Z. and Wessel, G. (2018) Isolation and molecular identification of lactic acid bacteria using 16s rRNA genes from fermented Teff (Eragrostis tef (Zucc.)) dough. Int J Food Sci. 2018, 8510620. Doi: 10.1155/2018/8510620
Zhu, Y., Zhang, Y., and Li, Y. (2009) Understanding the industrial application potential of lactic acid bacteria through genomics. Appl. Microbiol. Biotechnol Vol. 83 No 4: Pages 597-610