Department of Zoology, The University of Burdwan, Burdwan, 713104, India
Corresponding author Email: soumen.microbiology@gmail.com
Article Publishing History
Received: 02/01/2019
Accepted After Revision: 28/03/2019
The starch hydrolysing bacteria have important role in plant nutrient uptake. The aim of the present study is to isolate and characterize bacteria that are able to degrade starch from the rhizospheres of maize plants. Isolation and characterization were performed in the laboratory. The bacterial isolate (PCS1) was non-pathogenic and found to be positive for catalase, lipid hydrolysis and nitrogen fixation. The PCS1 also fermented sucrose, xylose, dextrose, galactose and mannose. The PCS1 strain showed halo-zone on starch medium and starch degrading index was found to be 2.14. The 16S rRNA gene sequence and phylogenetic tree analysis revealed that the PCS1 belonged to one of the strains of Burkholderia cepacia complex.
Amylase; Phylogenetic Analysis; Rhizosphere; Starch Degrading Index
Chatterjee P, Roy P, Mandal P, Chatterjee S. Isolation, Phenotypic and Molecular Characterization of Burkholderia Sp. (Strain, PCS1) from Maize Fields Exhibiting Starch Hydrolysis Ability. Biosc.Biotech.Res.Comm. 2019;12 (1).
Chatterjee P, Roy P, Mandal P, Chatterjee S. Isolation, Phenotypic and Molecular Characterization of Burkholderia Sp. (Strain, PCS1) from Maize Fields Exhibiting Starch Hydrolysis Ability. Biosc.Biotech.Res.Comm. 2019;12 (1). Available from: https://bit.ly/2JO8VEj
Copyright © Chatterjee et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
Introduction
A wide range of ecological niches have been occupied by the members of the genus Burkholderia sp. ranging from soils to the respiratory tract of humans (van Elsas et al., 2002; Coenye and Vandamme, 2003; Salles et al., 2002, 2004; Janssen, 2006). Beside free-living rhizospheric association with plants, some of the Burkholderia strains and species are also epiphytic and endophytic, including obligate endosymbionts and phytopathogen (Coenye and Vandamme, 2003; Janssen, 2006). The research studies on maize rhizosphere have shown that the Burkholderia are the predominant bacterial groups in maize of Italian agricultural fi elds (Hebbar et al., 1992; Ramette et al., 2005). In addition, Viallard et al. (1998) have shown that the species B. graminis and B. cepacia genomovar III are abundant in maize fi elds of France. The most abundant isolates belonged to the genus Burkholderia. Silva et al. (2016) studied the soils of the Amazon basin and found that Burkholderia spp. associated with the maize plants.
carry nifH genes and able to synthesize indole-3-acetic acid (IAA) and solubilize calcium phosphate. Recently, Correa-Galeote (2018) have shown that the cultivation history is the main driver of endophytic colonization of Burkholderia spp. in maize plants Bevivino et al. (1998) evaluated the metabolic and molecular profi les for some traits associated with biocontrol and plant growth promoting (PGP) activity in Burkholderia cepacia population. They showed that the B. cepacia strains displayed a wide antibiosis against the phytopathogenic fungi. Further studies on maize rhizosphere have shown the association of other species such as Burkholderia unamae (Caballero-Mellado et al., 2004) and B. silvatlantica (Perin et al., 2006a). The Burkholderia vietnamiensis was first described in the rice rhizosphere in Vietnam (Tran Van et al., 1994; Gillis et al., 1995) along with other Burkholderia species from the rhizosphere of maize and coffee in Mexico (Estrada-de Los Santos et al., 2001).
Several novel diazotrophic bacterial species belonging to the genus Burkholderia that may form both epiphytic and endophytic populations with the maize roots are known to be associated with N2 fi xation such as B. vietnamiensis (Estrada-de Los Santos et al., 2001; Estrada et al., 2002), Burkholderia tropica (Reis et al., 2004) and Burkholderia cenocepacia (Balandreau et al., 2001). Kost et al. (2014) demonstrated oxalotrophy as one of the vital traits of the genus Burkholderia phytofi rmans. So far, the studies were focused on the diversity of Burkholderia spp. in maize soils, N2 fi xing ability and plant growth promotion traits. The starch solubilising ability of Burkholderia spp. from maize roots is still not reported from any part of India. The objective of the present research work is to identify and characterization of Burkholderia spp. associated with maize rhizosphere capable of starch solubilization.
Materials and Methods
The research work was carried out in the Crop Research and Seed Multiplication Farm (CRSMF) of the University of Burdwan, West Bengal, located in the eastern part of India (latitude, 87°50’37.35’ E and longitude, 23°15’7.29’N and average altitude of 30m above mean sea level) using one variety of Zea mays L. (var. RE-55, Royal England).The soil samples were collected at the depth of 10 cm using soil auger from the rhizosphere of maize plants in sterile polythene bags and were taken to the Microbiology and Parasitology Laboratory of the University of Burdwan for bacterial analysis. The soil was air-dried and diluted up to 10-3 using sterile distilled water. From this dilution 100μl portion was mixed with sterile nutrient agar medium (NA) (peptone: beef extract: NaCl: agar at 5:3:3:1 g l-1) and was distributed in 5 sterilized plates. The plates were incubated at 30 ± 0.1 ºC in the BOD incubator for 24 hours to obtain isolated colonies. The colonies were observed on the incubated plates and the most abundant colonies (S1) were purified on NA plates. The bacterial isolate was maintained on the NA slants at 4 ± 0.1º C for subsequent characterization and identification. The morphological characters such as shape, size, colour, margin and opacity of the isolated colonies were determined. The bacteria was streaked on different media including starch (Starch Agar, Himedia, India) and free-living nitrogen-fi xing media (Mannitol, 10 g l-1; K2HPO4, 0.5 g l-1; MgSO4. 7H2O, 0.2 g l-1; NaCl, 0.2 g l-1; MnSO4.4H2O, trace; FeCl3, trace; Agar powder 10 g l-1) and plates were kept at 30 ± 0.1 ºC in the BOD incubator.After 24 hours, standard microbiological methods were followed for morphological and biochemical characterization of the isolate (Holt et al., 1994; Sneath and Holt, 2001; Forbes et al., 2007).
The Gram staining was performed using Gram staining kit (Himedia, India). Motility test was done by stabbing the bacteria into SIM agar (Czaban et al., 2007). The biochemical properties such as catalase, citrate utilization, nitrate reduction, indole production, Methyl-Red (MR), Voges-Proskauer (VP), urease, oxidase, and carbohydrate metabolism (acid-gas production) tests were performed. The pathogenicity test was done on Blood Agar and by DNase test (Benson et al., 2012). Carbohydrate fermentation test was performed using the sugar discs of maltose, adonitol, fructose, inositol, mannose sucrose, raffi nose, dextrose, cellobiose, galactose, rhamnose, xylose, melibiose, lactose, salicin, ducitol, sorbitol, mannitol, trehalose and arabinose (Azmi et al., 2014).
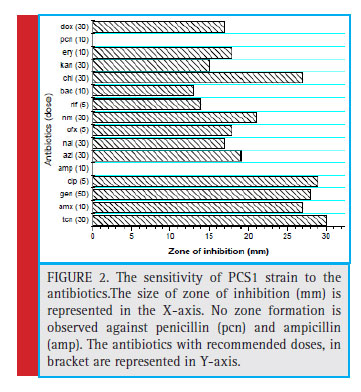
In addition, the qualitative determination of enzymes such as starch hydrolysis, lipid hydrolysis, protein hydrolysis (gelatine) tests were also performed. The starch plates were fl ooded with Grams Iodine after 24 hours of incubation. The ability of the bacteria to degrade starch was described by Starch Degrading Index (SDI), which is the ratio of the total diameter of clear zone and colony diameter (Nusrat and Rahman, 2007). The sensitivity analysis of the isolate to the recommended doses of antibiotics was done following Brown et al. (2004). The strain was tested for tetracycline (tcn), amoxicillin (amx), gentamycin (gen), ciprofl oxacin (cip), azithromycin (azi), nalidixic acid (nal), ofl oxacin (ofx), neomycin (nm), rifampicin (rif), bacitracin (bac), chloramphenicol (chl), kanamycin (kan), erythromycin (ery), ampicillin (amp), penicillin (pcn) and doxycycline (dox). The zone interpretation was done following CLSI (2011).
Genomic DNA isolation kit was used to isolate the genomic DNA of the bacterial strain from the pure culture pellet (DNA-Xpress TM Kit, Himedia-MB501). The rDNA fragment was amplifi ed by PCR and the product was sequenced by Genetic Analyzer (ABI 3130, Genetic Analyzer) bidirectionally using the forward and reverse primer. The bacterial 16S rDNA fragment was amplified with primers 8F (5’-AGA GTT TGA TCC TGG CTC AG-3’)
and 1492R (5’-ACG GCT ACC TTG TTA CGA CTT-3’). The following PCR cycling conditions were used in the study: 94oC for 300 s; followed by 36 cycles of 94oC for 30 s, 58oC for 60 s, and 72oC for 60 s; and then a final elongation step at 72oC for 7 minutes.
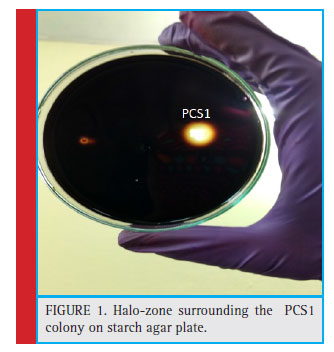 |
Figure 1: Halo-zone surrounding the PCS1 colony on starch agar plate. |
Results and Discussion
The colony of the bacterial isolate (PCS1) was white, round with an average diameter of 6mm, opaque and fl at (Fig. 1). The Gram staining of the bacteria showed rod-shaped and Gram-negative properties of this strain. The motility test of the strain indicated non-motile character. The bacterial isolate (PCS1) showed a positive result for catalase and negative for MR, VP, citrate reductase, nitrate reductase and indole production (Table 1). The Triple Sugar Iron (TSI) showed yellow slant and yellow butt, and also gas production. The pathogenicity test in Blood Agar and DNase agar showed no halo-zone formation. The carbohydrate fermentation results are shown in the Table 2. The strain (PCS1) was able to ferment sucrose, xylose, dextrose, galactose and mannose. The bacteria responded positively to the lipid hydrolysis and negatively to the protein (gelatine) hydrolysis test. The bacterial strain (PCS1) was able to grow on nitrogen-free medium.The starch hydrolysis test showed a positive result for the bacteria. The SDI calculated from the halo zone and colony ratio, which was found to be 2.14 in the present study.The bacterial strain was found to be sensitive to the recommended doses of the antibiotics used in the study except ampicillin and penicillin, which showed with no zone formation. The results of the zone of inhibition exhibited by the strain is shown in Fig. 2. Based on the morphological, biochemical and phylogenetic tree analysis, the PCS1 isolate was identifi ed as one of the strains of Burkholderia cepacia complex. The restriction map of the 16S rRNA gene sequence of PCS1is shown in Fig. 3. The molecu-lar percentage of GC and AT content were 59.20% and 40.80 % respectively. The phylogenetic tree showed that the PCS1 was branched with Burkholderia cepacia ATCC 55792 (AY741359) (Fig. 4).
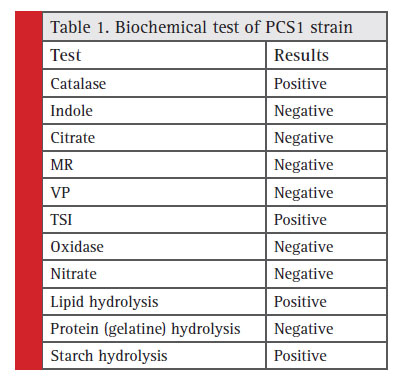 |
Table 1: Biochemical test of PCS1 strain |
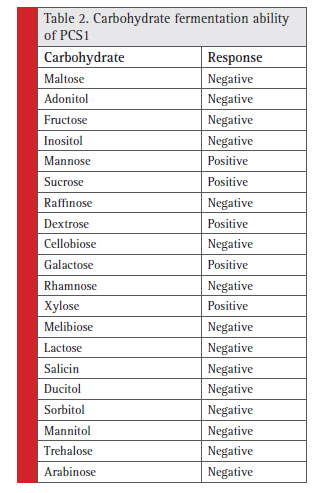 |
Table 2: Carbohydrate fermentation ability of PCS1 |
 |
Figure 3: The fi gure showing the restriction map of the 16S rRNA gene sequence of the isolate PCS1. |
 |
Figure 4: The neighbourhood joining tree of the PCS1 strain based on 16S rRNA gene sequence |
The bacterial communities are harboured by plant roots and shoots. In the rhizosphere, the root exudates are known to be the source of carbon and energy for the bacterial population. The secretion of root exudates varies with plant species and growth phases. These exudates are of highly diverse chemical nature ranging from small carboxylates to complex phenolic compounds. These exudates are utilized as substrates by the bacterial groups colonized in the rhizosphere. Brito et al. (2018) isolated Burkholderia ambifaria from the rhizoplane of the maize plants and showed that colonization of this bacterial isolate is responsible for an increase in shoot growth, root and root hair. The amylase is secreted extra-cellularly by starch degrading rhizobacteria and facilitate other different organic matters for plants to easily absorb and manufacture their food (Cordeiro et al., 2003).
Smits et al. (1996) advocated that the rhizobacteria often show the high value of the starch degrading index, which belongs to Gram-positive and spore-forming bacterial groups; and the enzyme production from the microorganism is directly correlated to the time period of incubation. Another fi nding on starch degradability by amylase was shown by Wang et al. (2016), the authors observed that the ability to hydrolyse starch by amylase is independent of the diameter of the colony formed by Gram-negative, non-spore forming rhizobacteria. The calculated SDI value of PCS1 is quite high in comparison to some other gram-negative bacteria.
The strains of Burkholderia spp. are capable of producing amylase enzymes to degrade starch into a soluble form and promote the maize plant growth. Dida (2018) studied the SDI of Bacillus spp. from the rhizosphere of various plants in Ethiopia and reported that the SDI falls between 1.23 and 2.15. The present fi ndings in relation to SDI is comparable with Bacillus spp.. Draghi et al. (2014) reported two strains of Burkholderia cordobensis namely LMG 27620T and LMG 27621 from Argentina. They observed that the strains were well capable of starch hydrolysis. The DNA G+C content of the strain PCS1 was 59.20 mol %, which is within the range reported for members of genus Burkholderia (59-69.9 mol %; Gillis et al., 1995). The present results are in agreement with the research works of Draghi et al. (2014) and Lim et al. (2012). The results of carbon compound utilization by the PCS1 strain of Burkholderia cepacia are corroborated with the findings of Yabuuchi et al. (1992).
Dalmastri et al. (1999) studied the effect of soil type, maize cultivar and root localization on the genetic diversity of Burkholderia cepacia populations and found that the degree of genetic diversity was regulated strictly by the soil types. Salles et al. (2004) added that the distribution of Burkholderia spp. in soil is attributed to the land use history of that area beside the maize cultivar and soil type. It is probably due to the soil type and maize cultivar, which were responsible for the deviation of the characteristics of the novel strain, PCS1 from the other strains of Burkholderia cepacia, reported from other parts of the world.
Conclusion
In addition to nitrogen fi xation in soils, Burkholderia spp. also exhibit starch degrading property that is beneficial to maize plants. The DNA G+C content and the carbohydrate fermentation abilities of the novel strain, PCS1 supports the properties of the genus Burkholderia. The phylogenetic tree showed the clustering of PCS1 with Burkholderia cepacia. It can be said that the variation of the strains from the other strains of Burkholderia were due to the type of soil and maize cultivar used in the study.
Acknowledgements
The fi rst author is thankful to the Department of Zoology, The University of Burdwan for giving all sorts of laboratory facilities to conduct this research. Thanks are due to the agricultural scientists of the Department of Agriculture, Govt. of West Bengal and scientists of CRSMF, The University of Burdwan for extending the help to carry out the research.
References
Azmi, S.A., Catterjee, S.N., Saha, N.C. (2014). Phenotypic and molecular characterization of bubble eye gold fi sh pathogenic ampicillin resistant Bacillus sp(GF1). Int. J. of Pharma. Bio. Sci., 4: 342-351.
Balandreau, J., Viallard, V., Cournoyer, B., Coenye, T., Laevens,S., Vandamme, P. (2001). Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol., 67: 982–985.
Benson, D.A., Karsch-Mizrachi, I., Clark, K., Lipman, D.J., Ostell, J., Sayers, E.W. (2012). GenBank. Nucleic Acids Res., 40: 48-53.
Bevivino, A., Dalmastri, C., Tabacchioni, S., Chiarini, L., Belli, M.L., Piana, S., Materazzo, A., Vandamme, P., Manno, G. (2002). Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol., 40: 846–851.
Brito, T.S., Buss, R.P.L.A., Carvalho, J.P.F.C., Eberling, T., Martinez, A.S., Vandeir Francisco Guimarães, V.F., Chaves, E.I.D.O. (2018). Growth promotion of Burkholderia ambifaria associated to nitrogen fertilization in the initial development of corn. J. Agric. Sci., 10(6): 123-135.
Brown, A. (2004). Benson’s Microbiological applications: Laboratory Manual in General Microbiology. McGraw-Hill Science/ Engineering/Math, New York.
Caballero-Mellado, J., Martinez-Aguilar, L., Paredes-Valdez, G., Estrada-de los Santos, P . (2004). Burkholderia unamae sp. nov, and N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol., 54: 1165–1172.
CLSI (2011). Clincal Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. Wayne, Pennsylvania, USA.
Coenye, T., Vandamme, P., Govan, J.R.W., Li Puma, J.J. (2001d). Taxonomy and identifi cation of the Burkholderia cepacia complex.
J. Clin. Microbiol., 39: 3427–3436.
Cordeiro, C.A.M., Martinas, M.L.L., Lucaino, A. (2003). Production and Properties of alpha amylase from thermophylic Bacillus
species. Brazil J. Microbiol., 33: 1-3.
Correa-Galeote, D., Bedmar, E.J., Arone, G.J. (2018). Maize endophytic bacterial diversity as affected by soil cultivation history. Front. in Microbiol., 9: 484.
Czaban, J., Gajda, A., Wroblewska, B. (2007). The motility of bacteria from rhizosphere and different zones of winter wheat roots. Polish. J. of Environ. Stud., 16 (2): 301-308.
Dalmastri, C., Chiarini, L., Cantale, C., Bevivino, A., Tabacchioni, S. (1999). Soil type and maize cultivar affect the genetic diversity of maize root-associated Burkholderia cepacia populations. Microb. Ecol., 38: 273-284.
Draghi, W.O., Peeters, C., Cnockaert, M., Snauwaert, C., Wall, L.G., Zorreguieta, A., Vandamme, P. (2014). Burkholderia cordobensis sp. nov., from agricultural soils. Int. J. System. Evol. Microbiol., 64: 2003-2008.
Estrada, L.S.P., Bustillos-Cristales, R., Caballero-Mellado, J. (2001). Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol., 67: 2790–2798.
Estrada, L.S.P., Mavingui, P., Cournoyer, B., Fontaine, F., Balandreau, J., Caballero-Mellado, J. (2002). A N2 -fixing endophytic Burkholderia sp. associated with maize plants cultivated in Mexico. Can. J. Microbiol., 48: 285–294.
Forbes, B.A., Sahm, D.F., Weissfeld, A.S. 2007. Bailey & Scott’s Diagnostic Microbiology, Elsevier, Inc., St. Louis, MO, USA, 12th edition.
Gillis, M., Van, T., Bardin, R., Goor, M., Hebbar, P., Willems, A., Segers, P., Kersters, K., Heulin, T., Fernandez, M.P. (1995). Polyphasic taxonomy in the genus Burkholderia leading to an amended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol., 4(5): 274–289.
Hebbar, P., Davey, A.G., Merrin, J., Dart, P.J. (1992). Rhizobacteria of maize antagonistic to Fusarium moniliforme , a soil borne fungal pathogen: colonisation of rhizosphere and roots. Soil Biol. Biochem., 24: 989–997.
Holt, J.G., Krieg, N.R., Sneath, P.H.A., Staley, J.T. and Williams, S.T. (1994). Bergey’s Manual of Determinative Bacteriology (9th Int. ed.), Williams and Wilkins, Maryland, USA, pp. 565- 596.
Janssen, P.H. (2006). Identifying the dominant soil bacteria taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol., 72: 1719–1728.
Jukes, T.H., Cantor, C.R. (1969). Evolution of protein molecules. In: Munro, H.N., (ed). Mammalian Protein Metabolism, New York. Academic Press.
Kost, T., Stopnisek, N., Agnoli, K., Eberl, L., Weisskopf, L. (2014). Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofi rmans. A Report, University of Zurich.
Krisle da Silva, K., Perin, L., Gomes, M.L., Barauna, A.C., Pereira, G.M.D., Mosqueria, C.A., Costa, I.B., O’hara, G., Zilli, J.E. (2016). Diversity and capacity to promote maize growth of bacteria isolated from the Amazon region. Act. Aamaz., 46(2): 111-118.
Lim, J.S., Choi, B.S., Choi, A.Y., Kim, K.D., Kim, D.I., Choi, I.Y., Ka, J.O. (2012). Complete genome sequence of the fenitrothiondegrading
Burkholderia sp. strain YI23. J. Bacteriol., 194: 896.
Nusrat, A., Rahman, S.R. (2007). Comparative studies on the production of extracellular -amylase by three mesophilic Bacillus isolates. Bangladesh J. of Microbiol., 24(2): 129-132.
Perin, L., Martinez-Aguilar, L., Paredes-Valdez, G., Baldani, J.I., Estrada-de Los Santos, P., Reis, V.M., Caballero-Mellado, J. (2006a). Burkholderia silvatlantica sp. nov., a diazotrophic bacterium associated with sugar cane and maize. Int. J. Syst. Evol. Microbiol., 56: 1931–1937.
Ramette, A., LiPuma, J.J., Tiedje, J.M. (2005). Species abundance and diversity of Burkholderia complex in the environment. Appl. Environ. Microbiol., 71: 1193–1201.
Reis, V.M., Estrada-de los Santos, P., Tenorio-Salgado, S. (2004). Burkholderia tropica sp. nov, a novel nitrogen-fixing, plant associated bacterium. Int. J. Syst. Evol. Microbiol., 54: 2155–2162.
Saitou, N., Nei, M. (I987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol., 4: 406-425.
Salles, J.F., De Souza, F.A., van Elsas, J.D. (2002). Molecular method to assess the diversit y of Burkholderia species in environmental
samples. Appl. Environ. Microbiol., 68: 1595–1603.
Salles, J.F., van Veen, J.A., van Elsas, J.D. (2004). Multivariate analyses of Burkholderia species in soil: effect of crop and land use history. Appl. Environ. Microbiol., 70: 4012–4020.
Smits, J.P., Rinzema, J., Tramper, H., Van, M., Knol, W. (1996). Solid-state fermentation of wheat bran by Trichoderma reesei QM9414: substrate composition changes, C balance, enzyme production, growth and kinetics. Appl. Micro. Biotechnol., 46:489-496.
Sneath, P.H.A., Holt. J.G. (2001). Bergey’s Manual of Systematic Bacteriology (2nd ed.), Williams & Wilkins, Springer-Verlag, NY, USA, 1, pp. 64.
Tran Van, V., Mavingui, P., Berge, O., Balandreau, J., Heulin, T. (1994). Promotion de croissance du riz inocule par une bacteria fixatrice d’azote, Burkholderia vietnamiensis , isolee d’un solsulfat e acide du Vietnam Agronomie., 14 : 697–707.
Van Elsas, J.D., Garbeva, P. Salles, J. (2002). Effects of agronomical measures on the microbial diversity of soils as related to the suppression of soil-borne plant pathogens. Biodegradation, 13: 29–40.
Viallard, V., Poirier, I., Cournoyer, B., Haurat, J., Wiebkin, S., Ophel-Keller, K., Balandreau, J. (1998). Burkholderia graminis sp. nov.,a rhizospheric Burkholderia species, and reassessment of Pseudomonas phenazinium ,Pseudomonas pyrrocinia and Pseudomonas glathei as Burkholderia. Int. J. Syst. Bacteriol., 48: 549–563.
Wang, S., Lin, C., Liu, Y., Shen, Z., Jeyaseelan, J., Qin, W. (2016). Characterization of a starch-hydrolyzing -amylase produced by Aspergillus niger WLB42 mutated by ethyl methane sulfonate treatment. Int. J. Biochem. Mol. B., 7(1 ): 1-10.
Yabuuchi, E., Kosako, Y., Oyaizu, H., Yano, I., Hotta, H., Hashimoto, Y., Ezaki, T. & Arakawa, M. (1992). Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol., 36: 1251–1275.