Department of Biological Sciences, Faculity of Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
Corresponding author email : sdjastaniah@kau.edu.sa
Article Publishing History
Received: 04/12/2020
Accepted After Revision: 20/03/2021
Bacterial resistances to the existing antibiotics are increased and multidrug resistant bacteria pose a serious threat worldwide which cause morbidity and mortality throughout the world. Thus, new antimicrobial agents are needed. The present study was focused on isolation of Actinomycetes with abilities to produce diffusible pigments in media from different parts of the East side of the Umm Jirsan cave, Saudi Arabia. Samples were collected at different depth of the soil surface and different distances from the cave entrance (30-550 m).Among 107 isolates, a total of 66 isolates were diffusible pigment actinomycete producers. Among the 66 pigment producing Actinomycetes, 12 isolates were selected based on the intensity and color of the pigment.
These isolates were screened for their antimicrobial activities against some tested microbial pathogen by different methods, cross streak method, Agar plug diffusion method and Agar well diffusion methods. Strain SAG-85 was the most active isolate. The identification and biochemical characterization of the isolate SAG-85 were determined. It was belonging to genus Streptomyces. Furthermore, the extracellular pigment was extracted using ethyl acetate and this extract showed excellent antibacterial activities against Serratia marcescens, and MRSA but very weak effect on Staphylococcus aureus. In conclusion, Streptomyces species played a critical role as a source of pigments with antibacterial activities.
Actinomycetes, Umm Jirsan Cave, Antibacterial Activity, Microbial Pathogen, MRSA
Al-Ghamdi S. A, Jastaniah S. D, Amasha R. H. Isolation and Screening of Actinomycetes from Umm Jirsan Cave, Saudi Arabia for their Antibacterial Activity. Biosc.Biotech.Res.Comm. 2021;14(1).
Al-Ghamdi S. A, Jastaniah S. D, Amasha R. H. Isolation and Screening of Actinomycetes from Umm Jirsan Cave, Saudi Arabia for their Antibacterial Activity. Biosc.Biotech.Res.Comm. 2021;14(1).. Available from: <a href=”https://bit.ly/3reV7DI“>https://bit.ly/3reV7DI</a>
Copyright © Al-Ghamdi et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Caves are one of the harsh environments on earth, unique in nature, unexploited, and poorly studied (Cheeptham et al., 2013).They can be classified based on type of rock and formation method. The most common types of caves are limestone and other calcareous rocks (Northup and Lavoie, 2001).Microbial diversity of caves attract the attention of many microbiologists (Barton, 2006) and microorganisms obtained from unexplored caves may belongs to new taxa which produce unique or novel bioactive compounds that are important to human (Genilloud, 2017; Kemung et al., 2018; Takahashi and Nakashima,2018).
On other hand, unexplored environment such as Cave have a significant potential for exploring new antimicrobial substance because of the increased competition in the environment due to the limited nutrients content, as result of which the microorganism produces antimicrobial substance against each other. For example, Streptomyces, which was isolated from a volcanic cave, has a high yield of secondary metabolites (Cheeptham et al., 2013). Novel antimicrobial materials from new actinomycetes, isolated from Kazakhstan desert (extreme habitats) were studied and approximately half of screened isolates inhibits microbial growth (Ziyat et al. 2019).
They added that Kazakhstan soils are rich reservoirs of new actinobacteria with antimicrobial activity. Streptomyces sp. SM01 from Indian soil produce novel antimicrobial agent, picolinamyc which inhibit S. aureus with the MIC value of 0.04-5.12µg/ml (Maiti et al., 2020). Seven isolates of Streptomyces, isolated from Iranian dry soil, exhibited antimicrobial activity against resistant and sensitive test organisms (Majidzadeh et al., 2021).
The unique characteristics of cave like reduction of light, high humidity with acidic, low nutrients and lack of oxygen (Schabereiter et al., 2002) which encourage bioactive substances production by the found bacteria (Nakaew et al., 2009). These compounds mostly displayed anti-bacterial and/or anti-cancer activities. Two major groups of soil Actinomycetes are Streptomyces and Micromonospora (Arifuzzaman et al., 2010). Streptomyces are Gram positive, aerobic spore formers and possess DNA rich in GC content (69-73 %), filamentous and they form extensive branching substrate and aerial mycelia (Khamna et al., 2010).
Actinomycete produces many kinds of bioactive compounds and they also have excellent ability to produce natural pigments (Parmar et al., 2016). A total of 22,500 bioactive secondary metabolites have been reported, out of which 16,500 compounds show antibiotic activities. Moreover, out of the 22,500 total bioactive secondary metabolites, 38% from fungi, 17% from other bacteria (Berdy, 2005), 10,100 (45%) are reported to be produced by actinomycetes in which 7630 from Streptomycetes and 2470 from rare Actinomycetes. Species of Streptomyces, account for more than 70% of the total antibiotic production (Lam, 2006) and have long been recognized as prolific producers of useful bioactive compounds (Majidzadeh et al., 2021).
Many kinds of antibiotics are produced by actinomycetes and moreover of these antibiotics contain pigments which are usually described in different colors such as blue, violet, red, rose, yellow, green, brown and black. The pigments may be dissolved into the medium or retained in the mycelia (Amsaveni et al., 2015).They played an important role in characterization of these organisms and are widely used as coloring agents in industries such as, dyeing industry, printing industry, food industry, textile industry, pharmaceutical industry and cosmetic production (Lee et al., 2006).
Streptomyces coelicolor and S. violaceoruber produce an important red-blue antibiotic actinorhodin and associated compounds like α-, β-, ε-, γ-actinorhodin. These pigments have a wide range of applications in scientific, medical and industrial sector. So, it is a beneficial alternative (Palanichamy et al., 2011). Streptomyces hygroscopicus (strain D10) from desert soil produce extracellular yellow pigment which showed good activity against some pathogens (Selvameenal et al., 2009).
Microbial biopigments are safe and are alternative to synthetic dyes which have potential health hazard and cause diseases like cancers, allergies. Currently, the whole world is looking towards the usage of natural pigments over synthetic colorants. According to the World Health Organization (WHO) over prescription and the improper use antibiotics has led to the generation of antibiotic resistance in many bacterial pathogen (Tandal et al., 2018).
Rangseekaew and Pathom-aree (2019) selected 47 species in 30 genera of actinobacteria from cave and cave related habitats. Several novel actinobacterial taxa were isolated from caves habitats during the period of 20 years. The highest number of novel species was from genus Streptomyces followed by Amycolatopsis and Nocardia. Actinomycetes achieve excellent roles in production of secondary metabolites.
Most of the antibiotics in use today are natural secondary products of bacteria, Actinomycetes and fungi. The present study was focused on the isolation of some Actinomycetes producing pigments from Umm Jirsan cave and screening them for their antibacterial activities against some bacterial pathogens.
MATERIAL AND METHODS
General discretion of the studied area: In 2007, Umm Jirsan cave in Saudi Arabia was considered as one of the longest lava tube system in Saudi Arabia and it was founded to be ~1481.2 meter long, located in near the center of Harrat Khaybar Lava Field ,130 km north of Al-Madinah which is in western part of the Kingdom of Saudi Arabia (Figure 1).
Figure 1: A:Location of Harrat Khaybar Lava Field in Al-Madinah, Saudi Arabia, B:Details of map showing the two entrances located at east and west sides of the cave (Al-shanti and Pint, 2007).
The location (39°,41° west and 25°,26° North) was visited during the study period with the help of a local cave explorer (Dr. Mahmoud Ahmed Al-shanti, Saudi Geological Survey).The average temperature was approximately 30˚C in October 2018. The system consists of three lava-tube passages separated by two collapses height of 8-12 m and a maximum passage width of 45 m (Figure 1).
Sediment covering the cave floor was measured at 1.17 m deep. The cave is about 2050 meters above sea level in the volcanic region to the east and 320 m in the valley area to the west (Pint, 2009). The cave has two opening, the entrance at the West side and the entrance of the East passage (Figure 2).
Figure 2: A:Location of Umm Jirsan cave in Harrat Khyber, B:The entrance of West side, C: The entrance of East passage.

Collection of soil samples: Cave soil samples were collected from different parts of the East side of Umm Jirsan lava cave tube system (Figure 2). From ether 5 or 20 cm depth of surface and different distances from the cave entrance (30 m, 200m , 250 m, 350 m, 450 m, and 550 m also from the exit of the cave.
The samples were collected in a sterilized containers and kept under 4ºC for 24 hrs until transferred to laboratory, then some of the samples were spread on Starch nitrate agar (SNA) medium and some were suspended in sterile water and serial dilutions are made and the suitable dilution was used for actinomycete isolation.
Isolation and purification of Actinomycetes: The isolation of actinomycetes was done by standard serial dilution method (Valan et al., 2009). One gram of soil samples was mixed in distilled sterile water up to 10-3 and allowed for shaking with vortex for 5 minutes and 0.1 ml of sample from dilution were inoculated on SNA using streak plate technique. Plates were incubated at 30oC for 5-7 days. All the pigments producing Actinomycetes that grown on plates were picked and purified by streak plate method (Katz, 2008).
Tested pathogenic microbes: Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Enterococcus faecalis, Serratia marcescens and Salmonella sp. were obtained from King Fahad hospital in Jeddah, Saudi Arabia while Methicillin-resistant Staph. aureus (MRSA) was from Eye Hospital, Jeddah, Saudi Arabia.
Preparation of the bacterial suspension: In sterile Falcon tube, 5ml of sterile broth nutrient broth medium was inoculated by fresh pathogenic bacteria culture. All tubes were incubated at 37˚C for 24 hrs and growth was measured by the optical density (OD)of the suspension at 540 nm using Spectrophotometer. The OD of each suspension was adjusted to 0.5-0.6 McFarland turbidity standards (Mcfarland,1907).
Screening of pigments producing Actinomycetes for antimicrobial activity: All the pigmented isolates of Actinomycetes from the cave soil sample were screened for inhibitory substances production against microbial pathogens.
Primary screening using Cross streak method: The cross streak method (Oskay, 2009) is applied by inoculating a single streak of 12 selected isolates on the center of the Mueller Hinton agar plate and incubated for 5 days at 30ºC, after incubation, the plates are seeded with test pathogenic microorganisms in a perpendicular arrangement around the Actinomycetes strains and finally the plates were incubated for 24 hrs at 37˚C. The microbial interaction was analyzed by determining the distance of inhibition measured in mm, isolates showing highest inhibition zone were secondary screened.
Agar plugs diffusion method: The isolate SAG-85 was cultured on SNA medium and incubated at 30˚C for 5 days. After incubation, the medium was cut aseptically with sterile corkborer with 8 mm diameter and deposited on opposite side on MHA plate, instantly inoculated with 0.1 ml of suspensions of pathogenic microbes. All plates were incubated for 24hrs at 37˚C. The secondary substances diffuse from the plug to MHA medium. The antimicrobial activity of the isolates was detected by the appearance of inhibition zone which measured by mm around the agar plug.
Agar well diffusion method: A disc (8 mm diameter) from the pigmentedSAG-85 isolate was grown on SNA medium at 30˚C for 5-7 days, then transferred to 250 ml conical flasks containing 48 ml SN broth medium. The flasks were incubated in incubator shaker (120rpm) at 30˚C for 5 days to obtain the extracellular pigments. After incubation, SNB cultures were centrifuged at 4,500 rpm for 30 min.
the supernatant was collected and filtrated through micro filter 0.2 µm to remove cells. Then, well with diameter 6 mm punched aseptically by a sterile corkborer on MH agar plates inoculated by the test microbial pathogen. Then, 100 µl of each supernatant was loaded into each well and the plates were left for one hour on refrigerator at 4˚C. Then, the plates were incubated for 24 hrs at 37˚C. The antimicrobial agent diffuses in agar medium and inhabits the growth of the tested pathogenic bacteria. The mean diameter of the produced inhibition zone was determined.
Extraction of pigments from the selected isolate: After incubation of the selected isolate SAG-85 for 5days at 30˚C, the broth was centrifuged at 4,500 rpm for 15 min and the supernatant was collected and filtrated through microfilter 0.2 µm. Pigment extractions from culture was extracted as described by Naikpatil and Rathod (2011).
Ethyl acetate was added to culture filtrate in equal volumes (1:1,v/v) and shaken vigorously overnight in incubator shaker (120rpm) at 30˚C.Then, the mixture of Ethyl acetate and filtrate were added in separating funnel, shacked well for 15 min and left for 1 hr. This step was repeated twice and the upper aqueous layer containing the bioactive pigment was collected and evaporated in fume hood to dryness. After evaporation, the concentrate of Ethyl acetate was dissolved in methanol and transferred into sterilized glass tube and stored at 4˚C for antimicrobial assay.
Antimicrobial activity assay after extraction of the bioactive secondary metabolite: The antimicrobial activity of bioactive extract for isolate SAG-85 was assayed by agar well diffusion method. This method is widely used to evaluate the antimicrobial activity of plants and microbial extract (Valgas et al., 2007, AL-Ansari, et al., 2019)
Four wells of 6 mm diameter were obtained by a sterile corkborer dug on MHA plates which was inoculated with 0.1 ml, contained 4×106 cfu/ml of the bacterial suspensions of S. marcescens, Staph. aureus and MRSA. After sterilization by micro filter (pore size 0.2 µm), 100µl of the extract was loaded to each agar well and methanol was tested as control. All the plates were left for one hour in refrigerator at 4˚C. Then, incubated for 24hrs at 37˚C.The antimicrobial activity of bioactive compound or extract was determined by measuring the mean area ofthe inhibition zone around the well in mm.
Characterization of Actinomycetes: The morphological characterization and biochemical tests based on Bergey’s Manual of Systematic Bacteriology were carried out for the most active bacterial isolate (Holt et al., 1989).
Morphological characterization of the selected isolate: Morphological characterization of the selected isolate of actinobacteria was described on SNA medium, different agar media and under light microscope.
Light microscopic examination: Examination under light microscope using oil immersion lens for the most active bacterial isolate was carried out after Gram staining.
Actinobacteria growth and description on different agar media: Growth on different media was detected by the International Streptomyces Project (I.S.P) introduced by Shirling and Gottlieb (1966). The media used were tryptone-yeast extract (ISP-1), yeast- malt extract agar (ISP-2), oatmeal agar (ISP-3), inorganic salt-starch agar (ISP-4), glycerol-asparagine agar (ISP-5), tyrosine agar (ISP-7) and carbon, nitrogen utilization medium (ISP-9) and SNA. Culture was incubated for 5-7 days at 30˚C. The degree of growth (poor , moderate, heavy or no growth) and the colony colors of the aerial and substrate mycelia and diffusible pigments were described (Vishwanatha et al., 2017)
Biochemical test of selected isolate: The selected isolate SAG-85 was biochemically characterized by API-20E test. Catalase test, oxidase test, starch hydrolysis, coagulate test, blood hemolytic, melanin pigments production, antibiotics sensitivity were also recorded.
Molecular identification of selected isolate: The selected isolate SAG-85 was grown on SNA medium for five days at 30˚C,then, transmitted to Macrogen for identification by 16S rRNA. The information of the used primer was summarized in (Table 1).
Table 1. Primers used for molecular identification of the isolate SAG-85.
| Primer Information | |
| Sequencing ( primer name and sequence) | PCR (primer name and sequence) |
| 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′ | 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3 |
| 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′ | 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′ |
RESULTS AND DISCUSSION
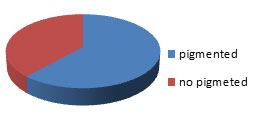
The cave soil samples were collected form East side of Umm Jirsan lava cave tube system, Harrat Khyber, Al-Madinah city, Saudi Arabia. Serially dilution soil samples produce wide range of diffusible pigment and non-diffusible pigment producing actinomycetes. Among 107 isolates, a total of 66 isolates (61.68 %) had diffusible pigment producing actinomycetes. About 41 (38.32%)isolates were non diffusible pigment producing actinomycetes (Figure 3).
Figure 3: The percentage of pigmented Actinomycetes obtained from the studied cave.
Among the 66 pigments producing actinomycetes (Figure4), 12 isolates were selected and screened for production of diffusible pigment on SNA. The selection of the isolate was based on intensity of the pigment and the strength of the color. The pigment colors were pink, yellow, brown, black and orange (Table 2). The 12 isolates were then screened for antimicrobial activity against tested microbial pathogen by cross streak method (Figure 5). The selected 12 actinomycetes isolates were subjected to secondary screening process.
Three isolates designated SAG-23, SAG-24, and SAG-85 showed a higher inhibition zone against all the tested microbial pathogen. Also, the isolates SAG-23, SAG-24 and SAG-85 gave the highest inhibition zone and selected for secondary screening which was carried by agar well and agar disc diffusion methods. Moreover, isolate SAG-85 showed excellent activities against E. coli, Staph. aureus and K. pneumoniae with inhibition zone ranged from 25 -35 mm. thus, it was selected for detail studies (Table 3, Figure 6).
Figure 4: The different colors of the pigmented actinomycetes obtained from the studied cave
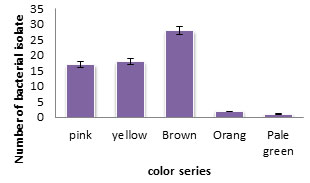
Table 2. Colony morphology of the selected pigment producing actinomycetes obtained from soil of Umm Jirsan cave samples at different distance on SNA medium.
| Isolates | Arial Mycelium | Substrate Mycelium | Diffusible Pigment | |
| 200 m | SAG-8 | White | Pink | Pink |
| SAG-15 | White | Dark pink | Pink | |
| SAG-23 | Purple | Dark pink | Pink | |
| SAG-24 | white | Dark pink | Dark pink | |
| 450-550 m | SAG-41 | Gray | Brown | Brown |
| SAG-48 | White | Black | Brown | |
| SAG-49 | Gray | Brown | Brown | |
| SAG-57 | White | Black | Yellow | |
| SAG-63 | White | Yellow | Dark yellow | |
| SAG-82 | White | Black | Brown | |
| SAG-85 | Gray | Brown | Yellow | |
| SAG-88 | pink | Brown | Brown |
Table 3. The antagonism effect between the selected isolates and some bacteria determined by inhibition zone measured in mm.
| Tested Bacteria | SAG-23 mm | SAG-24 mm | SAG-85 mm | Ampicillin(Control)
(5 µg/ml) |
| S. marcescens | 10.5 | 16.5 | 10.0 | 33.0 |
| P. aeruginosa | 10.5 | 14.5 | 10.0 | 22.0 |
| Staph. aureus | 25.5 | 25.5 | 35.0 | 34.0 |
| E. faecalis | 13.5 | 20.5 | 10.0 | 31.0 |
| K . pneumoniae | 10.0 | 10.0 | 31.0 | 25.0 |
| E. coli | 15.5 | 12.5 | 25.0 | 22.0 |
| Salmonella sp. | 18.0 | 16.0 | 10.0 | 22.0 |
| Staph. aureus (MRSA) | 12 | ND | 11 | 14.0 |
Figure 5: The Microbial antagonism between isolate SAG- 85and the tested bacteria
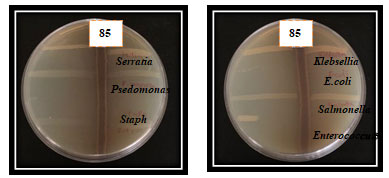
Figure 6: The inhibitory effect of the isolate SAG- 85 against different bacterial pathogens using agar well and agar disc diffusion methods.
Morphology characterization of the selected isolate: Under light microscope, the selected isolate SAG-85 appeared as Gram positive filamentous bacterium with aerial and substrate mycelia. On SNA medium, it had a heavy growth with gray color (Figure 7). The selected bacterium was grown on different agar media and the degree of growth and the colony colors of aerial and substrate mycelia and diffusible pigments were described in (Figure 8).
The conidia are in chain and the spore surface was smooth. The results of the physiological and biochemical tests of the selected isolate SAG-85 were shown in Table 4. The isolate showed positive results for Voges-Proskauer, Gelatinase, Coagulase, Amylase, Chitinase, Urease and Pectinase. The results of antibiotics susceptibility for the selected isolate SAG-85 and the utilization of different sugars were determined in Table 5.
Figure 7: The surface view (A), the bottom view (B)of the isolate SAG-85 grown on agar medium and the Gram stain of the selected isolate (C).
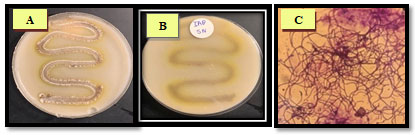
Figure 8: The growth of the isolate SAG-85 on diffrant agar media, (A): SNA, (B): ISP-1,(C):ISP-2, (D): ISP-3, (E): ISP-4 (F): ISP-5. (G): ISP-7 and (H): ISP-9
Table 4. biochemical tests of the selected isolate SAG-85
| Chemical test | SAG-85 | Chemical test | SAG-85 |
| o-nitrophenyl-β-D-galactopyranoside | + | Voges-Proskauer | + |
| Arginine dihydrolase | + | Indole Test | – |
| lysine decarboxylase | + | Gelatinase | + |
| Ornithine decarboxylase | + | Blood hemolytic | Alpha
hemolytic |
| Citrate | + | Oxidase | – |
| Hydrogen sulfide | + | Coagulase | + |
| Urease | + | Catalase | – |
| Tryptophan deaminase | – | Amylase | + |
| Chitinase | + | Pectinase | + |
| +: positive, -: negative | |||
Table 5. Antibiotic sensitivity tests and fermentation of sugar tests for the selected isolate SAG-85
| Antibiotics | Results | Sugar | Results |
| Ceftazidime (CAZ) | R | Arabinose | + |
| Aztreonam (ATM) | R | Mannose | + |
| Piperacilln (PRL) | R | Inositol | + |
| Imipenem (IMI) | S | Sorbitol | + |
| Ciprofloacin (CIP) | S | Rhamnose | + |
| Amikacin (AK) | S | Sucrose | + |
| R: Resistant, S: Sensitive, +: fermented | |||
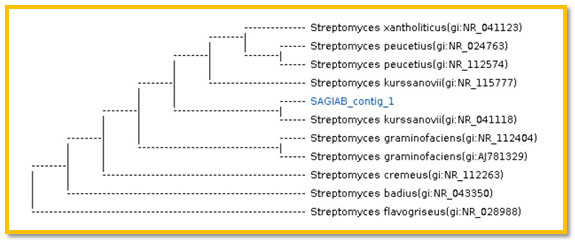
3.2 Molecular identification of selected isolate SAG-85: A phylogenic analysis was performed to identify the selected isolateSAG-85 using partial sequence of the 16S rRNA. The 16S rRNA sequence of the isolate SAG-85 showed high level of sequence similarity with member genus Streptomyces with homology level of 99% to Streptomyces kurssanovii as in the phylogenic analysis (Figure 9).
Figure 9: The phylogenic tree of the isolate SAG-85 (AB) and the most related isolates
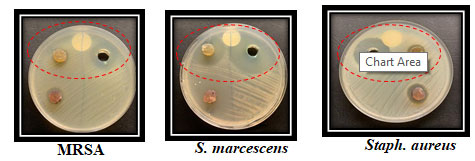
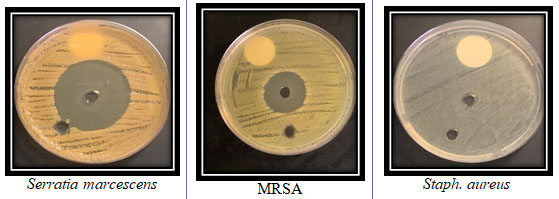
The supernatant containing the bioactive components were extracted with equal volume of ethyl acetate. The antimicrobial activity of the crude pigment extract of the Streptomyces kurssanovii was shown in (Figure 10).The highest inhibition zone was obtained against Serratia marcescens and MRSA with mean inhibition zone diameters of 47±2.4and 23±2.1mm, respectively. Very small inhibition zone was recorded for the tested extract against Staph. aureus (Figure 10).
Figure 10: The antimicrobial activty of theisolate SAG-85 organic extract against three bacterial pathogens
Cave environments are underexplored microbiologically where actinomycetes specially streptomycetes, have long been recognized as prolific producers of useful bioactive compounds with widely applications (Watve et al., 2001).The search for novel antibiotics and other bioactive microbial metabolites is important for the fight against new and emerging pathogens (Berdy, 2005, Busti et al., 2006). Isolation of actinomycetes from unique natural habitat is of interest to known bioactive metabolites. Streptomyces species still promise to remain fruitful sources of new antibiotics (Amin et al., 2016).
In the present study, 107 actinomycete isolates have recovered from cave soil sample from East side of Umm Jirsan lava cave tube, Harrat Khyber, Al-Madinah, Saudi Arabia. Out of 12 pigmented isolates, SAG-85 displayed marked inhibitory activity against bacterial pathogens in primary screening by cross streak technique.
The isolated actinomycetes generally showed slow growth and the cross streak method required a short incubation time so this method was difficult to obtain clear inhibition zone by cross streak method (Pereira and kamat, 2011) and some isolates showed no activity in primary and secondary screening (Dehnad et al., 2010).
Therefore, another screening method like agar plug method was used.Agar plug diffusion method is often used to highlight the antagonism between microorganisms and it is similar to disk diffusion method (Elleuch et al., 2010). The isolate SAG-85 was grown on SNA medium for 5 days at 30˚C and showed the highest antagonistic activity against MRSA, Staph. aureus, S. marcescens on in both plug diffusion method and agar well diffusion method.
Ethyl acetate was standardized as the best solvent to extract pigments (Selvameenal et al., 2009).The antimicrobial activity of the extract of the bioactive pigments of the isolate was active against all the tested bacterial pathogens. In this study the Actinomycetes have the ability to produce bioactive pigments with the antimicrobial activity and these materials are very useful for pharmaceuticals and other industrial aspects.
The most active isolate was characterized and identified as Streptomyces kurssanovii using 16S rRNA sequencing which is mainly used as a most powerful technique for bacterial identification (Yokota, 1997, XU et al., 1999). The 16S rRNA sequence was determined and phylogenic tree was obtained. Therefore, the Actinomycetes that have ability to produce natural bioactive pigment and has antimicrobial activity could be very useful for pharmaceutical and agricultural importance.
Similar to our isolate, Li et al., (2002) isolated Streptomyces scopiformis A25T from rhizosphere soil of China. This isolate was aerobic, mesospheric and with aerial and substrate mycelia. The spore chains are arranged in a broom -like structure arising directly from the substrate mycelium. Non fragmenting substrate mycelium consists of septet hyphae are extensively branched, rectiflexibles chains of roundish, spiny-surfaced spores. Moreover, Streptomyces kurssanovii was isolated from soil sample from Russia. Previous studies also revealed that aquatic and terrestrial environments are rich with Streptomyces isolates that are responsible for the production of various secondary metabolites.
In this study, the crude extract of the selected Streptomyces showed high activity against Gram-positive and Gram-negative bacteria. This means that the environmental factors may influence on the production of secondary metabolites. Recently, some Streptomyces isolates recorded excellent activity against the resistant isolates of MRSA. Species of the genus Streptomyces may produce glycopeptide materials that had high molecular weights (Zhu et al., 2013, Park et al., 2014, Tan et al., 2015, Al-Ansari et al., 2019, Majidzadeh et al., 2021).
CONCLUSION
Screening of a variety of actinomycetes for making of novel medicine is a needed practice. These new materials can be used against antibiotic resistant pathogenic bacteria. Extreme habitats like caves are rich sources of Actinomycetes which are proved to be important sources of novel useful antibiotic derivatives and new metabolites. Hence, the findings of this study revealed that Streptomyces sp. with antibiotic substances production capability was an important application. Also, Streptomyces species from unexplored regions are likely to yield novel antibacterial agents. It is very clear that Streptomyces sp. could be a promising microorganism for the development of novel antibacterial drug against a wide range of pathogenic bacteria
REFERENCES
Al-Ansari M., Alkubaisi N., Vijayaragavan P. and Murugan K. (2019) Antimicrobial potential of Streptomyces sp. to the Gram positive and Gram negative pathogens Journal of Infection and Public Health, V12, pp: 861–866.
Al-shanti M A and Pint J. (2007). Umm Jirsan Lava cave tube system, Harrat Khyber, Al-Madinah, maps of caves surveyed by Saudi Cave Unit in Saudi Geological Survey, Kingdom of Saudi Arabia: Saudi Geological Survey Data-File Report SGS-DF.
Amin S. M., Risan M. H., and Abdulmohimin N. (2016). Antimicrobial and Antioxidant Activities of Biologically Active Extract from Locally Isolated Actinomycetes in Garmian Area. Journal of Garmian University, V1(10) pp: 625-639.
Amsaveni R., Sureshkumar M., Vivekanandhan G., et al., (2015). Screening and isolation of pigment producing Actinomycetes from soil samples. international journal of bioscience an Nanosciences V2(2) pp: 24-28.
Arifuzzaman M., Khatun, M.R. and Rahman H.(2010). Isolation and screening of actinomycetes sundarbans soil for antibacterial activity. African Journal of Biotechnology, V9, pp: 4615-4619.
Barton A.H.(2006). Introduction to cave microbiology: A review for the nonspecialist. Journal of Cave and Karst Studies., V68, pp: 43-54.
Berdy J. (2005). Bioactive microbial metabolites. The Journal of Antibiotics, V58, P: 1-26
Busti E., Monciardini P., Cavaletti L.,Bamonte R., Lazzarini A., Sosio M. and Donadio S. (2006). Antibiotic producing ability by representatives of a newly discovered lineage of actinomycetes. Microbiology, V152, P: 675-683.
Cheeptham N., Sadoway T., Rule D., Watson K., Moote P., Soliman L.C., et al. (2013). Cure from the cave: volcanic cave actinomycetes and their potential in drug discovery. International Journal of Speleology. V42(1), P: 35–47.
Dehnad A.R.,Yeganeh L.P., Bakhshi, Soofiani S.A., Mokhtarzadeh A., et al., (2010). Investigation antibacterial activity of Streptomycetes isolates from soil samples, West of Iran. African Journal of Microbiology Research, V4 (14):1542-1549.
Elleuch L., Shaaban M., Smaoui S., et al., (2010). Bioactive secondary metabolites from anew terrestrial Sterptomyces sp. TN262, Applied Biochemistry and Biotechnology, V162 (2),P: 5779-593.
Genilloud O., (2017). Actinomycetes: still a source of novel antibiotics. Natural Product Reports. V34 (10), P: 1203–1232.
Holt J.G. and William S.T. (1989). Bergey Manual of systematic Bacteriology,V4.Lippincott Williams and Wilkins
Katz D.S. (2008). The Streak Plate Protocol-(online). Available from www.asmscience.org/content/education/protocol/protocol.3160.
Kemung H. M., Tan L. T. H., Khan T. M., Chan K. G., Pusparajah P., Goh B. H., et al., (2018). Streptomyces as a prominent resource of future anti-MRSA drugs. Frontiers in Microbiology, V9, pp: 1-26
Khamna S, Yokota A., Peberdy JF, and Lumyong S. (2010). Indole3-acetic acid production by Streptomyces sp. isolated from some Thai medicinal plant rhizosphere soils. Eur Asian Journal of Bio Sciences, V4, P: 23-32.
Lam K.S. (2006). Discovery of novel metabolites from marine actinomycetes. Current Opinion in Microbiology; V9 (3) pp: 245-251.
Lee C.L., Wang J.J., Kuo S.L. and Pan T.M. (2006). Effect of red mold rice on antifatigue and exercise-related changes in lipid peroxidation in endurance exercise. Applied Microbiol Biotechnology, V72, pp:1254-1262.
Li W., Lanoot B., ZhangY., VancanneytM., SwingsJ., Liu Z. (2002). Streptomyces scopiformis sp. nov., a novel streptomycete with fastigiate spore chains. International Journal of Systematic and Evolutionary Microbiolog, V52, pp: 1629–1633
Maiti P.K., Das S., Sahoo P. and Mandal S. (2020). Streptomyces sp. SM01 isolated from Indian soil produces a novel antibiotic picolinamycin effective against multi drug resistant bacterial strains. Scientific Reports, V10, pp1123-33.
Majidzadeh M., Heidarieh, P., Fatahi‑Bafghi, M. et al., (2021). Antimicrobial activity of Actinobacteria isolated from dry land soil in Yazd. Iran. Molecular Biology Reports, V 48 (1) pp: 121-133.
Selvameenal L. Radhakrishnan M. and Balagurunathan R. (2009). Antibiotic pigment from desert soil Actinomycetes: Biological activity, purification and chemical screening. Indian Journal of Pharmaceutical Sciences, V71, pp: 499-504.
McFarland J. (1907). Nephelometer: an instrument for media used for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. Journal of the American Medical Association, V14, pp:1176-8.
NaikpatilS.V. and Rathod J.L. (2011). Selective isolation and antimicrobial activity of rare actinomycetes from mangrove sediment of Karwar. Journal of Ecobiotechnology, V3, pp: 48– 53.
Nakaew N., Pathom-aree W. and Lumyong S. (2009). First record of the isolation, identification and biological activity of a new strain of Spirillospora albida from Thai cave soils. Society for Actinomycetes Japan, Actinomycetologica V23, pp: 1–7.
Northup D.E. and Lavoie K. H. (2001). Geomicrobiology of caves: a review. Geomicrobiol. J., V18, pp: 199-222.
Oskay M. (2009). Antifungal and antibacterial compounds fromStreptomyces strains. African Journal of Biotechnology,V8 (13) pp:3007-3017.
Palanichamy L., Hundet A, Mitra B. and Reddy N. (2011). Optimization of cultivation parameters for growth and pigment production by Streptomyces spp. isolated from marine sediment and rhizosphere soil. International Journal of Plant, Animal and Environmental Sciences, V1 (3), pp: 158-170.
Park HB, Lee JK, Lee KR. and Kwon HC. (2014). Angumycinones A and B, two new angucyclic quinones from Streptomyces sp. KMC004 isolated from acidic mine drainage. Tetrahedron V55, pp:63–6.
Parmar R.S., Singh, C., Jadon, P., Bhadauriya, G. and Kumar, A. (2016). Exploration of pigment producing Actinomycetes, isolated from Madhya Pradesh region of India. International Journal of Higher Education, V6 (2) pp:961-966.
Pereira S.V. and KamatN.M. (2011), Antimicrobial Screening of Actinobacteria using a Modified Cross- Streak Method.Indian Journal of Phamaceutical Sciences.V73 (2), pp: 223-228.
Pint J. (2009). Umm Jirsan: Arabia’s Longest Lava-tube System. Paper accepted for presentation at the 15th International Congress of Speleology,Kerrville, Texas,V2, pp: 714-717.
Rangseekaew P. and Pathom-aree W. (2019). Cave Actinobacteria as producers of bioactive metabolites. Frontiers in Microbiology V10, pp:387.
Schabereiter-Gurtner C., Saiz-Jimenez„ C, Pinar G., Lubitz W. and Röllke S. (2002). Altamira cave paleolithic paintings harbor partly unknown bacterial communities. FEMS. Microbiology Letters, V211, pp: 7–11.
Shirling ET and Gottlieb D. (1966). Methods for characterization of Streptomyces species. International Journal systematic bacteriology ; V16, pp:313-340.
Takahashi Y., Nakashima T. (2018). Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics V7 (3) pp: 1-2
Tan L.T., Ser H.L., Yin W.F., Chan K.G., Lee L.H, Goh B.H. (2015). Investigation of antioxidative and anticancer potentials of Streptomyces sp. MUM256 isolated from Malaysia mangrove soil. Frontiers in Microbiology, V6, pp:1316-22.
Tandal A., Khandagale M., Palaskar R. and Kulkarni S. (2018). isolation of pigment producing Actinomycetes from soli and screening their antimicrobial activity against different microbial isolates. International Journal of Current Research in Life Sciences, V7 (6), pp: 2397-2402
Valan Arasu M., Duraipandiyan V., Agastian P. and Ignacimuthu S. (2009). In vitro antimicrobial activity of Streptomyces sp. ERI-3 isolated from Western Ghats rock soil (India). Journal of Medical Mycology, V19 (1), pp:22-28.
Valgas C., De Souza S.M., Smania E.F.A. et al.,( 2007). Screening method to determine antibacterial activity of natural products. Brazilian Journal of Microbiology, V38, pp:369-380.
Vishwanatha B.T., Girish B. K., Padmashri S. B., Chethan J. D. and Sreenivasa N. (2017). Isolation, Identification and Characterization of Streptomyces sp. SN-2, Biosciences Biotechnology Research Asia, V14 (4), pp: 1401-1407
Watve M.G., Tickoo R., Jog M.M. and Bhole B.D.( 2001). How many antibiotics are produced by the genus Streptomyces, Archives of Microbiology, V176, pp: 386-390.
Xu L.H., Jin, X., Mao P.M., Lu Z.F., Cui X.L. and Jiang C.L.(1999). Three new species of the genus Actinobispora of the family Pseudonocardiaceae, Actinobispora alaniniohila sp. Nov., Actinobispora xinjiangensis sp. Nov. International Journal of Systematic Bacteriology, V49, pp: 88 1-886.
Yokota A. (1997). Phylogenic relationship of actinomyces. Atlas of actinomyces, Asakura Publishing Co. Ltd., Japan, pp:194-196.
Zhu H, Zhou X, Lei X.and Zhou Q. (2013) Antagonist of MRSA by Streptomyces sp. SJY056 from soil of the three-river headwater region in China Journal of Food Agriculture and Environment, V1(1), pp:420-2.
Ziyat A., Goddfellow M., Nurgozhina A., Sergazy S. and Nurgaziev M. (2019). Novel actinobacterial diversity in Kazakhstan deserts soils as a source of new drug leads. Journal of Microbiology, Biotechnology and Food Sciences,V8 (4) pp: 1057-1065.


