Shobhit Institute of Engineering & Life Sciences, Meerut, Uttar Pradesh, India
Corresponding author email: amarprakashgarg@yahoo.com
Article Publishing History
Received: 15/01/2022
Accepted After Revision: 30/03/2022
Human Faeces (HF) is a solid waste material that is secreted as left amount after digestion of food inside the small intestine of the body. It contains large number of viruses, bacteria, fungi, actinomycetes, archaebacteria etc of which Lactic Acid Bacteria (LAB) are very common and play an important role in digestion and immunity. Hence, LAB were isolated and enumerated from HFusing standard protocol in the present study to find out the qualitative and quantitative distribution of LAB in gut microbiome. LAB were isolated using MRS agar medium under anaerobic conditions and was found that Lactobacillus lactis, L. Acidophilus, L. fermentum and Enterococcus faecium were the dominant species and the populations varied from 3.5 x 106 to 4.5 x 1010CFU/mL. It shows that good populations of LAB in gut microbiome survive under anaerobic conditions. LAB have great efficiency to resist against antibiotics. Such species of LAB should be commercialized and marketed at a global stage so that problems related to imbalance in gut microbiome can be solved.
Anaerobicut Microbiome, Human faeces, Probiotics
Siddique S. P, Garg A. P. Isolation, Characterization and Quantitative Enumeration of Lactic Acid Bacteria from Human Faeces. Biosc.Biotech.Res.Comm. 2022;15(1).
Siddique S. P, Garg A. P. Isolation, Characterization and Quantitative Enumeration of
Lactic Acid Bacteria from Human Faeces. Biosc.Biotech.Res.Comm. 2022;15(1). Available from: <a href=”https://bit.ly/35CJz8K”>https://bit.ly/35CJz8K</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Keeping in view the health benefits of probiotic bacteria, several scientists are actively engaged in inventing/improving new strains of promoting gut bacteria, (Kechagia 2013; Ding, 2019).The microflora of gut get is involved in many biochemical processes and has various applications in human life. Previous studies have shown that maximum bacteria, isolated from the milk, were LAB (Jin2011; Dunlop and May 2015). It is a large group of bacteria used throughout the globe as probiotics. The group includes the microbes of common genera Lactobacillus, Lactococcus, Aerococcus, Enterococcus, Pediococcus, Leuconostoc, Streptococcus, Sporo lactobacillus, Vagococcus and Carnobacterium. Hence, LAB strains are bestowed with boons for infants and also for the adults (Bisht and Garg 2019; Cunningham 2021).
It has been observed that increased interest in probiotics in public, government organizations like Ministry of Food, Agriculture and Health, WHO and FAO and the industries dealing with medicines and healthy food, have led to greater focus on research on gut microbiome and LAB. Probiotics helps in enhancing the health by helping in digestion of food and maintains pH of the digestive system, production of useful products that helps in eliminating the bad microbes(Amaraand Shibl 2015).It is well known that 70% of the immunity is controlled by gut microbiome.It is also known that the efficiency and safeguard of various pathologies, gastrointestinal disorders, several allergies causing diseases, acute diarrhoea along with necrotizing enterocolitis help paediatric healthcare professionals with the help of probiotics strains(Martinelli et al. 2020).
The developments that are going with respect to science of microbiome are enabling the innovative research in the field of prebiotics and probiotics. The applications of probiotics and prebiotics have the capability to enhance the understanding as well as healthcare applications.Mixture of different LABs living in host gut provide huge health benefits (Swanson 2020).The probiotics are not only restricted up to the gut associated diseases alone, but also help in management of various acute and chronic disorders. This is because of the fact, that says probiotics are capable of modifying the microbial ecosystem of the intestine, increase the resistance functionality of the gut, provide anti-microbial substances, provide adherence to the mucosa and epithelium, enhance immunity response whether it is innate or acquired, (Nazi2018; Michaelet al. 2020; Cunningham 2021).
It was reviewed the powerful preventive and therapeutic function of probiotics for high serum cholesterol, allergic and HIV diseases, cancer and providing possible action mechanisms, (Nazi2018).Gut microbiome is responsible for the digestion of food material and the fibres are digested to produce short chain fatty acids like butyric acid that cause happiness and can help in the treatment of depression too (Duchmann1999;Gregoire 2020; Michaelet al. 2020).In an investigation, it was confirmed that a great diversity of microbial populations is present within the saliva and human gut.
Since, these microbes are of human origin, they may exhibit maximum functionality in the drugs and food which are there for human consumption. The living microbial organisms obtained from cultured milk and fermented foods are used for making foods for infants, also called as health friendly bacteria, which shows several health beneficial characteristics like for intolerance due to lactose, bowel diseases prevention, immune network system improvement, balance among the intestinal microbes, showing anti-hypertensive and anti-hypercholesterolemic, postmenopausal disorder alleviation, helps reducing traveller’s diarrhoea etc (Balakrishnan2016;Hao et al.2018; Bazireh2020).
The work from our laboratories has demonstrated the human colostrum’s is rich in LABs and their populations are greatly influenced with the diet of pregnant mothers (Arya 2020).At present, the most importantly used food agents are probiotics and prebiotics, that in combination act as symbiotic organisms. The dairy food products are regarded as the healthy product. Probiotic foods beneficially affect the host improving the life and induction of living microbial dietary agents in gastrointestinal flora, that regulate and stimulate the growth and development and the activation of catabolic mechanism of one or several health promoting bacteria in intestinal tract(Nagpal and Kumar 2012; Kailyn2020).
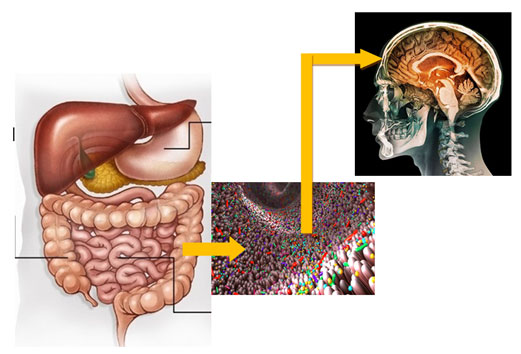
Recent evidences suggest that gut microbiome contains more than 3000 different microbes, however, only few have been cultured till date.These microbes give the signal to the brain which controls all live activities of the body. Vedic teachings that Jaisa Anna Vaisa Man seems to be based on scientific principles because the gut microflora is controlled with the type and quality of the food and the thought process activates in the brain is under the control of gut microbiome which is depicted in Fig. 1 (Kelly, 2015; Evelyn 2019; Maria 2020).
Trillions of microbes have evolved with and continue to live on human beings. With the rapid advances in tools and technology in recent years, new knowledge and insight in cross-talk between the microbes and their hosts have gained. The role of microbiota and mechanisms involved in the progress and development of major human diseases, that include obesity, hypertension, cardiovascular disease, diabetes, cancer, Inflammatory Bowel Disease (IBD), gout, depression have been reviewed in previous studies (Ding 2019; Maria 2020).
The human microbiome comprises bacteria, archaea, viruses and eukaryotes which reside within and outside our body. These organisms impact human physiology, both in health and in disease, contributing to the enhancement or impairment of metabolic and immune functions. Microorganismsare known to colonise various sites on and in the body (Ogunrinola and John 2020).Gut microbes affect the physiology of their hosts. Studying their diversity and functions is thus one of utmost importance as it will open new avenues towards the discovery of new biomolecules and the treatment of diseases.
Gut microbiome research is currently boosted by the unification of metagenomics, which has dominated the field in the last two decades, and cultivation, which is experiencing a renaissance (Thomas2021). Sehgal and Andreasson (2020) have found that there is bidirectional gut microbiota–brain communication in mood disorders. Effects of probiotics on brain connectivity and mental health outcomes and pregnancy related stress on gut microbiota in the newborn child has also been studied and positive relationship has been indicated (Sehgal and Andreasson 2020; Thomas 2021).
Figure 1: Transfer of signals from Gut Microbiome to Brain
The samples of Human Faeces (HF) were collected from 15different individual volunteers aseptically and were brought to the laboratory immediately in the School of Biological Engineering and Life Sciences, Department of Biotechnology, Shobhit Institute of Engineering and Technology, Meerut (UP) India. The individuals were told about the entire project and their consent was obtained. For the isolation of LAB, approximate 1 g of faeces samplewas quickly dissolved in 99mL of sterile distilled water from which dilution series (up to 10-8) was made. 0.1 mL suspension from dilutions10-6,10-7and 10-8were inoculated aseptically onto MRS agar plates by spread plate method. The plates were incubated at 37±1°C for 48 to 72h under anaerobic conditions using anaerobic gas jar. The plates were observed for bacterial colonies after incubation period, their morphology, color and texture were noted down carefully and colony forming unit per mL were counted using following formula.
Number of colonies x dilution Factor
CFU/ml = ———————————————————-
Volume Plated
For gram staining, smear was prepared on plain glass slide using a drop of saline and small amount of fresh bacterial culture from the plate. The smear was heat fixed gently and carefully, stained as per standard Gram’s stain method and was observed under oil emulsion lens 10 x 100x of compound light microscope (Carl Zeiss Microscopy GmbH). As LAB are Gram positive in nature, all the isolates which showed purple (positive) color were further processed for physiological and biochemical tests (Arya 2020).For catalase test, the 3% hydrogen peroxide solution was mixed gently over the surface of clean glass slide containing test microbial culture and was observed for formation of bubbles. Positive reaction was indicated by the presence of bubbles (Kumar and Kumar 2015, Asto et al. 2019). As LAB are catalase negative, all isolates that showed negative results were further tested for oxidase test.
For oxidase test, Cytochrome C oxidase test was performed using a filter paper soaked with the freshly prepared solution of tetramethyl-p-phenyl diamine dihydrochloride over which fresh culture of test bacteria was gently rubbed using sterile nichrome wire loop. Change in color within 30 second showed positive reaction. (Kumari 2008). As LAB are oxidase negative. Sugar Fermentation tests were performed for each isolate to confirm their Genus and species as per the recommendations of Bergey’s Manual of Systematic Bacteriology (9th edition).Disk containing 25 mg amount different sugars such as Maltose, Glucose, Lactose, Galactose, Mannitol, Xylose, Fructose was placed in 10mL of distilled water added and Durham tube was placed in inverted position. These were then autoclaved at 15Psi for 15 min andafter autoclaved 1 mL of inoculums of test isolate was added aseptically. These were incubated and observed for acid and gas production. These isolates were categorized on the basis of fermentation group (Koll 2010).The results are presented in (Table 2) and were interpreted as using Bergey’s Manual of Systematic Bacteriology.
RESULTS AND DISCUSSION
All 15 HF samples examined in our study, showed positive growth of LAB on MRS agar plates. The characteristics of colonies formed on plates varied in the form, shape, size and colour (Table 1) and ranged between 3.5x 106 to4.5x 1010..Sample no HF 7 showing the lowest number of colonies belonged to a person who was weaker and had suffered from gastric problem. HF10, HF15, HF11, HF5 and HF1 were collected from a healthy person taking good nutritious vegetable diet. In our preliminary studies, we have found that the diet seems to play significant role. Further experiments are being carried out in our laboratory with controlled diet.All the isolates tested in our study were found Gram’s positive and negative for catalase and oxidase which suggested that the isolated colonies belonged to the lactic acid bacteria (LAB) group. Based on morphological characters, texture, color of the colony and physiological and biochemical tests including sugar fermentation coupled with gas production, we identified the isolated species as Lactobacillus lactis, L. acidophilus, L. fermentumand Enterococcus faecium (Fig.2; Table 2)as per the recommendations of Bergey’s Manual of Systematic bacteriology.
The microphotographs of the pure culture of identified species are shown in Fig 3.The cells of L. lactis were spherical to ovoid and found in pairs or short chains, these were Gram’s positive and did not form spores.L. acidophilus showed rod shaped cells, Gram positive and fermented lactose. L. fermentum fermented glucose, lactose, galactose, fructose and sucrose (Table 2) and was Gram’s positive rods while Enterococcus faecium fermented maltose, lactose, mannose, galactose, fructose and sucrose and showed Gram’s positive reaction with rod shaped cells. It was found that Lactobacillus acidophilus was most common in all 15 samples and was detected in 80% of the samples followed by L. lactis, Enterococcus faecium and L. fermentum (Fig. 2, Table 2) on MRS agar medium under anaerobic/partially aerobic conditions. Almedia (2021)have characterized functional and taxonomic groups of human gut microbiota and have presented a Unified Human Gastrointestinal Genome (UHGG) collection comprising 204,938 non-redundant genomes from 4,644 prokaryotes which encode >170 million proteins (Almedia2021).
Roughly, 40 trillion microbes, consisting of more than 3000 species of several viruses, bacteria, archaea, fungi and other eukaryotes that live in and on our body and are extremely important for our health, however, few cause diseases and disorders (Wang 2018). Microbial biomass in/on our body consists of 1 Kg (approximately) representing about 1000,000 genes while entire human genome consists of approximate 30,000 genes and the human microbial genome greatly influence the physiology and behaviour of human. Gut microbiome is the principal ecological niche and secretes various enzymes that are mainly responsible for the digestion of food and also accounts for 70% of the total immunity of the body (Stefan 2020). Lactic acid bacteria (LAB) constitute the major part of gut microbiome but these are difficult to culture because of the requirement of anaerobic / partially aerobic conditions for their culture. Most scientists have focussed on genomic characterization of gut microbiota and a large number of microbial species have been identified (Samuel 2019; Stefan 2020).
However, very few species have been cultured so far. The isolation, characterization and culture of 4 species of LAB from faeces as representative of gut microbiota of human in this paper suggest that gut microbes if cultured under anaerobic / partially aerobic conditions on MRS agar media, can help in isolation of more and more species which can then be evaluated for their commercial exploitation. Diet influences the gut microbiome and the importance of plant-based diet in regulation of gut microbiome and its impact on human brain functions has been reviewed by (Medawar 2019; Angelis 2020). Gut bacteria can also help to manage depression (Pichler 2020; Chevalier 2020). Extraction of probiotic Lactobacillus and Enterococcus strains from faeces and human saliva was also reported in previos studies (Bazireh 2020).It is suggested that human gut microbiome need to be cultured so that their probiotic value may be evaluated and used for commercial purposes(Bazireh 2020).
Figure 2: Isolated LAB on MRS agar plates
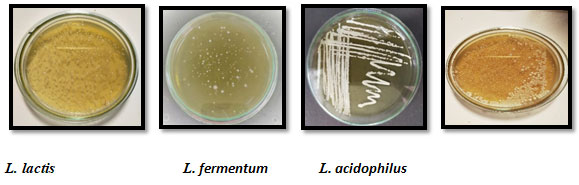
Figure 3: Microphotographs of isolated strains of LAB under 10 x 100X (original magnification)
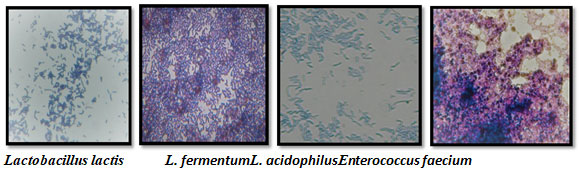
Table 1. Morphological characteristics and quantitative number of isolated LAB.
| Sample No. | Isolates
No |
Colour | Shape | Size | Margin | Opacity | Elevation | Texture | CFU/ mL |
| HF 1 | SUB-101 | Pale Yellow | Circular | Small
|
Entire | Opaque | Raised | Smooth | 2.8 x 1010 |
| HF 2 | SUB-102 | Off white | Circular | Moderate | Entire | Transparent | Flat | Moist | 3.5 x 107 |
| HF 3 | SUB-103 | Creamy white | Circular | Large | Entire | Opaque | Convex | Dry | 2.5x 107 |
| HF 4 | SUB-104 | Bright white | Circular | Small | Entire | Opaque | Flat | Powdery | 3.9 x 1010 |
| HF 5 | SUB-105 | Pale yellow | Circular | Moderate | Entire | Transparent | Raised | Smooth | 4.6x 109 |
| HF 6 | SUB-106 | Creamy white | Circular | Small | Entire | Transparent | Flat | Moist | 4.1x 108 |
| HF 7 | SUB-107 | Pale yellow | Circular | Moderate | Entire | Opaque | Convex | Smooth | 3.5x 106 |
| HF 8 | SUB-108 | Off white | Circular | Large | Entire | Opaque | Flat | Dry | 2.6x 108 |
| HF 9 | SUB-109 | Pale yellow | Circular | Large | Entire | Transparent | Raised | Moist | 2.9 x 1010 |
| HF10 | SUB-110 | Creamy white | Circular | Small | Entire | Opaque | Convex | Dry | 4.5x 1010 |
| HF11 | SUB-111 | Off white | Circular | Moderate | Entire | Transparent | Flat | Smooth | 6.3 x 109 |
| HF12 | SUB-112 | Creamy white | Circular | Small | Entire | Opaque | Convex | Dry | 4.6 x 108 |
| HF13 | SUB-113 | Pale yellow | Circular | Small | Entire | Opaque | Raised | Moist | 4.5x 107 |
| HF14 | SUB-114 | Creamy white | Circular | Small | Entire | Opaque | Flat | Smooth | 3.4 x 107 |
| HF15 | SUB-115 | Pale yellow | Circular | Small | Entire | Transparent | Convex | Dry | 4.8 x 109 |
Table 2. Characterization of different isolated strains based physiological and biochemical tests.
| Species isolated | % Occurrence
In 15 samples |
Gram’s staining | Catalase
test |
Oxidase
test |
Cell form | Fermented
Sugars (+) |
Unfermented Sugars
(-) |
| Lactobacillus acidophilus | 80.0% | + | – | – | Rod | Glu, Mal, lac, Gal | Man, Fru, Suc |
| L. lactis | 66.67% | + | – | – | Coccobacillus | Glu, Mal, Lac, Man | Gal, Fru, Suc |
| L. fermentum | 53.3% | + | – | – | Cocci | Glu, Mal, Fru, Suc | Lac, Man, Gal, |
| Enterococcus faecium | 60.0% | + | – | – | Rod | Mal, Gal, Fru | Glu,Lac, Man, Suc |
CONCLUSION
The findings of the present study suggest that the lactic acid bacteria can be isolated, characterized and cultured from human faeces as representative of gut microbiota on MRS agar plates under anaerobic/partially aerobic conditions. We have successfully isolated, characterized and cultured Lactobacillus lactis, Lactobacillus acidophilus, Enterococcus faecium and Lactobacillus fermentum from 15 human faeces samples.It may safely be concluded that probiotics play a vital role in the management of the health of human beings. Proper concentration and diversity of species of probiotics are necessary for the maintenance of the immunity of the ruminants.
We have isolated, cultured and characterized LAB from human faeces where we found that immediate processing of the samples was extremely essential. Most of the studies in western countries are confined to genomics and proteomics of gut microbiome while the culture of microbes is essential for their further commercial utilization. With this view point, our studies are very important and we suggest that the microbiologists working on gut microbiome should make all prior preparations for inoculation before sampling and the time between the sampling and inoculation should be less than 10 minutes (personal experience of our entire group).
ACKNOWLEDGEMENTS
The study was financially supported by the Shobhit Institute of Engineering & Technology, Meerut, Uttar Pradesh, India.
Conflict of interests: Authors have no conflict of interests to disclose.
Data Availability Statement: The database generated and /or analysed during the current study are not publicly available due to privacy, but are available from the corresponding author on reasonable request.
REFERENCES
Alexandre A.A., Nayfach S., Boland M. et al. (2021). A unified catalog of 204,938 reference genome from the human gut microbiome. Nature Biotechnology (23), 105-114.
Amara A.A. and Shibl A. (2015). Role of Probiotics in health improvement, infection control and disease treatment and management. Saudi Pharmaceuticals Journal. 23(2): 107-114.
Asto E, Mendez L, Audivert S, et al. (2019). The efficacy of probiotics, prebiotic Inulin- Type Fructans, and Synbiotics in Human Ulcerative Colitis: A Systematic Review and Meta- Analysis. Nutrients. 11,293.
Bazireh H., Shariati P., Jamalkandi A.S., et al. (2020). Isolation of Novel Probiotic Lactobacillus and Enterococcus Strains from Human Salivary and Fecal Sources. Frontiers in Microbiology. 11 Article 597946.
Brogan A.D.M.K., Ferrocino I., Calabrese F.M. et al. (2020). Diet influences the function of the human intestinal microbiome. Nature. 10:4247. doi/org/10.1038/s41598-020-61192-y.
Cunningham M., Azcarate-Peril M.A., Barnard A., et al. (2021). Shaping the Future of Probiotics and Prebiotics. Trends in Microbiology 29(8): 667-685.
Ding R., Goh W., Wu R. et al. (2019). Revist gut microbiota and its impact on human health and disease. Journal Of Food And Drug Analysis. 27:623-631.
Ding R.X., Goh W.R., Wu R.N., et al. (2019). Revisit gut microbiota and its impact on human health and disease. Journal of Food and Drug Analysis. 27: 623e631.
Duchmann R, May E, Heike M, et al. (1999). T cell specificity and cross reactivity towards Enterobacteria, Bacteroides, Bifidobacterium, and antigens from resident intestinal flora in humans. Gut. 44, 812–818. doi: 10.1136/gut.44.6.812
Dunlop A.L., Mulle J.G., Ferranti E.P., et al. (2015). Maternal microbiome and pregnancy outcomes that impact infant health: A Review. Adv. Neonatal Care. 15(6): 377-385.
Forster S.C., Kumar N., Blessing O.,et al. (2019). A human gut bacterial genome and culture collection for improved metagenomic analyses. Nature Biotechnology(37): 186-192.doi/org/10.1038/s41587-018-0009-7.
Grace A. Ogunrinola, Oshamika J.O.O. et al. (2020). The human microbiome and its impacts on health. Hindawi International Journal of Microbiology. 804-56-46.7.
Gregoire C., Siopi E. and Guenin-Mace L. (2020). Effect of gut microbiota on depressive-like behaviours in mice is mediated by the endocannabinoid system. Nature Communication doi.org/10.1038/s41467-020-19931-2.
Hao W., Wei C., Min L., et al. (2018). Good or bad: gut bacteria in human health and diseases. Biotechnology and Biotechnological equipment. 32(5): 1075-1080.doi.org/10.1080/13102818.2018.1481350.
Hirani, Komal J., Sandeep K. et al. (2021). Biochemical characterization and probiotic potential of Lactic Acid Bacteria Isolated from camel milk. Biosc. Biotech. Res. Comm.14(1): 196-202
Huhn M.E.S., Villringer A., Witte V. et al. (2019). The effects of plant- based diets on the body and the brain: a systematic review. Translational Psychiatry 9: 226.doi.org/10.1038/s41398-019-0552-0.
Jin L., Hinde K., Tao L. et al. (2011). Species diversity and relative abundance of lactic acid bacteria in the milk of rhesus monkeys (Macaca mulatta). J. Med. Primatol. 40(1): 52-58.
Kechagia M., Basoulis D., Konstantopoulou S., et al. (2013). Health Benefits of Probiotics: A Review. ISRN Nutrition. 2013/ Article ID 481651.1-7.
Koll P, Mandar R, Smitdt I, et al. (2010). Screening and evaluation of human intestinal lactobacilli for the development of novel gastrointestinal probiotics. Current Microbiol, 61:560-564.
Kumar A and Kumar D (2015). Characterization of Lactobacillus isolated from dairy samples for probiotic properties. Anaerobic 33:117-123.
Kumar A.R., Singh J., GargA.P. et al. (2020). Isolation, Characterization and Evaluation of Probiotic Potential of Lactic Acid Bacteria Isolated from Human Colostrum.Biosc. Biotech. Res. Comm.13(1):296-306
Kumari A., Garg A.P., Makeen K., Lal M. et al. (2008). A bacteriocin production on soya nutria nuggets extract medium by Lactococcus lactis subsp. Lactis CCSUB202. Int.J. dairy Sci. 3(1).49-54.
Martinelli M., Banderali G., Bobbio M et al. (2020). Probiotics’ efficacy in paediatric diseases: which is the evidence? A critical review on behalf of the Italian Society of Pediatrics. Italian Journal of Pediatrics. 46: 104. 1-13.
Nagpal R., Kumar A., Kumar M., et al. (2012). Probiotics, their health benefits and applications for developing healthier foods: a review. FEMS Microbiol Lett. 334: 1-15.
Nazir Y., Hussain S.A., Hamid A.A, et al. (2018). Probiotic and Their Potential Preventive and Therapeutic Role for Cancer, High Serum Cholesterol. BioMed Research International. 2018/ Article ID 3428437:1-17.
Neha B. and Garg A.P. (2019). Antagonistic activity of Lactic Acid Bacteria Against Common Enteric Pathogens Isolated from Milk and Milk products and Evaluation of their Probiotic Attributes. Bioscience Biotechnology Research Communications. 12(4). In Press.
Pichler M.J., Yamada C., Shuoker B. et al. (2020). Butyrate producing colonic clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nature Communication 11:3285|doi.org/10.1038/s41467-020-17075-x|.
Sehgal J., Ellionore and Andreasson A. (2020). The gut microbiota and mental health in adults. Current Opinion in Neurobiology, 62:102–114. https//doi.org/10.1016/j.conb.2020.01.016
Shi L.H., Balakrishnan K., Thiagarajah K., et al. (2016). Beneficial Properties of Probiotics. Tropical Life Sciences Research. 27(2): 73-90.
Stefan K.L., KimM.V., lwasaki A. et al. (2020). Commensal Microbiota Modulation of Natural Resistance to Virus Infection. Kasper. 10:047.
Swanson K.S., Gibson G.R., Hutkins R., et al. (2020). The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus statement on the definition and scope of synbiotics. Nature Reviews Gastroenterology & Hepatology. 17: 687-701.
Thomas C.A and Afrizal H. (2021). Recent advances in culture-based gut microbiome research. International Journal of Medical Microbiology. 311-151-485.


