1Department of Microbiology, University of Burdwan, Burdwan, West Bengal, India
2Department of Botany, Vivekananda Mahavidyalaya, Haripal, Hooghly, West Bengal, India.
Corresponding author email : sinhasangramvm@gmail.com
Article Publishing History
Received: 06/12/2020
Accepted After Revision: 22/03/2021
Ralstonia solanacearum is a devastating pathogenic soil borne bacterium causing Bacterial Wilt disease in 450 plant species belonging to 54 botanical families and it severely impairs global solanaceous crop production. The loss of crop may go up to 90% depending upon the environmental suitability. The bacterium is very robust and can survive in diverse host plants, soil, water and even in weeds. It possesses an arsenal of secretory molecules like diverse virulent factors, exopolysaccharide, cell wall degrading enzymes to subvert host defense mechanisms. The wilt pathogen is also very efficient to overcome existing control measures rendering it extremely difficult to control.
Understanding of molecular mechanism of pathogenesis through genome analysis and identification of novel drug target could be an effective alternative. In this study, subtractive genome analysis of Ralstonia solanacearum GM1000 strain having total 5106 proteins obtained from Uniprot database was done and 4972 non paralogous sequence of proteins were selected applying CD-HIT tool. A total of 465 essential proteins were then identified using BLASTp tools of DEG database. Functional pathway assessment of 424 essential proteins revealed 117 metabolically active proteins using KAAS server and a total of 7 non homologous proteins exclusive to the pathogen were identified using BLASTp algorithm.
After screening the druggability of 7 proteins in Drug Bank Database, 4 proteins were shortlisted and further analyzed for subcellular localization using PSORTb tool. After survey of the existing literature, type II secretory pathway gspe-related protein has been identified and predicted to be the best possible target for drug designing. The present work reports for the first time that type II secretory system could serve as drug target and therefore, opens a new avenue for in silico screening of novel molecules for effective control of bacterial wilt in future.
Drug Design, Ralstonia Solanacearum, Subtractive Genome Analysis, Wilt Disease.
Mandal J, Sinha S. In Silico Identification of Protein in Ralstonia solanacearum, A Bacterial Wilt Pathogen for Drug Target By Subtractive Genomic Analysis. Biosc.Biotech.Res.Comm. 2021;14(1).
Mandal J, Sinha S. In Silico Identification of Protein in Ralstonia solanacearum, A Bacterial Wilt Pathogen for Drug Target By Subtractive Genomic Analysis. Biosc.Biotech.Res.Comm. 2021;14(1). Available from: <a href=”https://bit.ly/36GJFcT”>https://bit.ly/36GJFcT</a>
Copyright © Mandal and Sinha This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Soil born bacterium Ralstonia solanacearum is the most devastating plant pathogenic bacteria that causes wilt diseases in many wide varieties of plants (Yuliar, Nion, and Toyota, 2015). The strains of this pathogen can infect 450 plant species distributed in 54 botanical families, including potatoes, tomatoes, brinjal, tobacco etc. (Wicker et al., 2007). It invades through the wounded roots or natural opening and colonize in the vascular tissues and release viscous exopolysaccharide that causes obstruction in xylem conduction and lead to fatal wilting disease symptoms in the plants (Schell MA, 2000). Direct yield losses by R. solanacearum vary widely according to the host, cultivar, climate, soil type, cropping pattern.
It has been reported that it accounts for 80% loss in tobacco, 100% in banana, and up to 20% in the groundnut (Elphinstone, 2005; Somani et al., 2010).Ralstonia infection causes more than 50% crop loss in India and that may reach up to 75% in some parts of Karnataka (Gadewar et al., 1991). Bacterial wilt disease affects potato cultivation in different parts of India and accounts for 30 to 70 % crop loss in these areas (Somani et al., 2010). The control of bacterial wilt pathogen is very challenging. Difficulties are associated with controlling this pathogen due to its abilities to grow endophytically, long survival in the soil especially in the deeper layers, travel along water, and its relationship with weeds (Wang et al., 2005; Mansfield et al., 2012 Santana et al., 2020; Yan and Gao, 2020).
The bacterial pathogen often undergoes VBNC (Viable but not culturable) state under unfavorable condition (Van Elsas et al., 2001). Furthermore, many environmental stresses weaken the defense systems of the plants allowing to proliferate Ralstonia and other bacterial endophytes inside the host. Conventional disease management practice such as preventive measures, cultural practices are inefficient to pre-existing infection and because of the pathogen’s diverse host range and persistence in the weeds and soil (Mbaka et al., 2013). Chemical pesticides such as algicide (3-[3-indolyl] butanoic acid), fumigants (Metam sodium, 1, 3-dichloropropene, and chloropicrin), and plant activators (validamycin A and validoxylamine) inducing systemic resistance in the tomato have been used to control bacterial wilt but with limited success (Ishikawa et al., 2007; Yuliar et al., 2015; Coutinho et al., 2017).
Copper compounds (copper hydroxide (CH), copper hydroxide-oxadixyl, and copper oxychloride-dithianon), and essential oils (Cinnamon oil, Clove oil) have been partially effective to control bacterial wilt. (Elphinstone, 2005; Lee et al., 2012). Many bactericides such as triazolothiadiazine [0.5 to 12 mM, in solution), streptomycin sulfate [400 mg kg−1 of soil] have been employed to control bacterial wilt pathogen with average rate of success (Khanum et al., 2005; Lin et al., 2010). Additionally, emergence antibiotic resistance and environmental pollution due to long-term use of chemical pesticides rendered bacterial wilt disease management very difficult. Although, there are many studies have been done employing biocontrol strategy to control bacterial wilt but of limited success due to inefficient colonization, narrow range and requirement of high inoculum of biocontrol agents.
Therefore, identification of novel pathogenic target protein and discovery of its corresponding drug could be an attractive alternative for controlling bacterial wilt disease (Whipps and Gerhardson, 2007; Coutinho et al., 2017).Rapid advancement in the field of biotechnology enabled us to have vast genomic data from the prokaryotic whole genome projects that in turn may be exploited for finding novel drug targets and virulent factors in microbes. With the availability of whole genome sequence, subtractive genome analysis has been evolved as a very efficient tool to identify novel drug targets and virulent factors in pathogenic microbes (Miesel et al., 2003; Amineni et al., 2010; Keshri et al., 2014).
Subtractive genome analysis is a smart technique to identify essential metabolic gene present exclusively in the pathogen having no homologue in the host and therefore, the targeted drug developed against the pathogenic essential metabolic gene will impair only the metabolic function of the pathogen leaving the host metabolism undisturbed (Vetrivel et al., 2011; Barh et al., 2011). Many possible drug targets have been identified in human pathogenic bacteria (Barh et al., 2011; Sudha et al., 2019; Santana et al., 2020; Yan and Gao, 2020).
However, there are very few reports regarding drug target identification in plant pathogenic bacteria using in silico techniques (Allen et al., 2009; Silver, 2011). Subtractive hybridization technique has been exploited to underpin drug targets in rice bacterial pathogen, Xanthomonas by some researchers (Keshri et al., 2014; Prava et al., 2019). Although, the complete genome sequence of Ralstonia solanacearum is available in the data base, but there is no report available so far that have tried subtractive genome analysis to identify drug targets in this bacterium. Therefore, the present work is attempted to identify possible drug targets in Ralstonia solanacearum through subtractive genome analysis and other in silico analysis tools (Prava et al., 2019).
MATERIAL AND METHODS
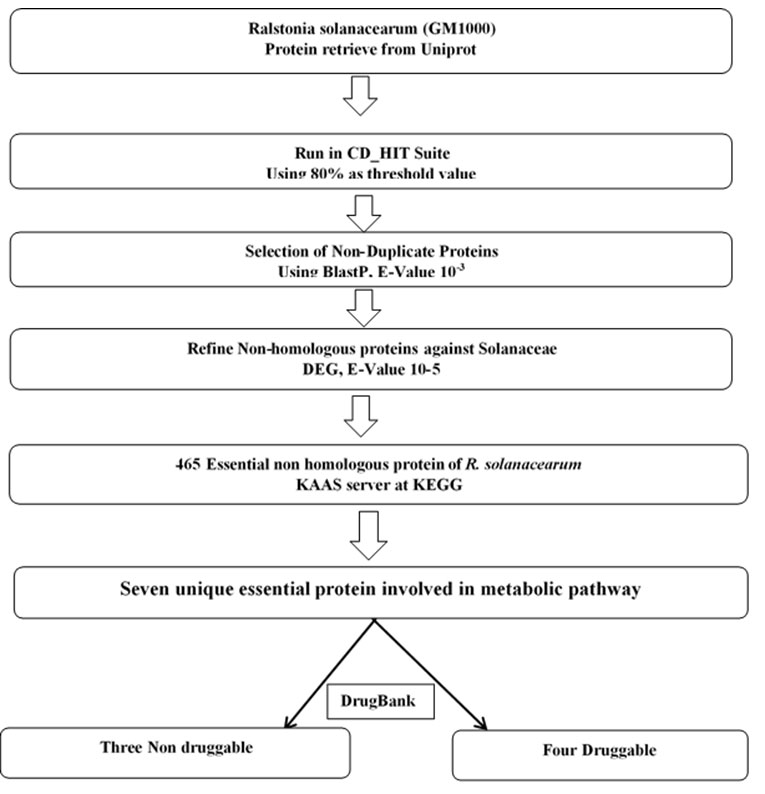
The conceptual framework showing the methodology followed for the analysis.followed for the analysis.Subtractive genomic approach was applied for the identification of essential proteins in the Ralstonia solanacearum (GM1000) which were then analyzed for the identification potential drug targets. The identified drug target was then screened through Drug Bank database to evaluate druggability scope. Network based analysis was done for the identification of metabolic activity of target protein (Yu et al., 2010).
The complete proteome of Ralstonia solanacearum GM1000 strain was retrieved from UniProt (http://www.uniprot.org). The UniProt Knowledgebase is the central hub for the collection of functional information on proteins, with accurate, consistent and rich annotation (The UniProt Consortium, 2019). Identification of nonhomologous protein and essential gene of Ralstonia solanacearum – Paralogous sequences were excluded from the complete proteome of Ralstonia solanacearum GM1000 strain by using CD-HIT at 80% threshold. BLASTp was performed for the remaining proteins against Solanaceae using threshold expectation value (E Value) 10-3 as parameter.
Non homologous protein sequences were then subjected to BLASTp against the database of essential genes (DEG) assessed at DEG database (http://tubic.tju.edu.cn/deg/) using E-Value cut off of 10-5, to screen out essential gene proteins (Li et al., 2001).KEGG Automatic annotation Server (KAAS) was accessed to analyze the metabolic pathway of the essential proteins of Ralstonia solanacearum GM1000 strain for the identification of potential drug target. The server performs BLASp comparisons of the query protein against Kyoto Encyclopedia of Genes and Genomes (KEGG) Genes Database (Moriya et al., 2007).
Sub Cellular localization of non-homologous essential proteins of bacteria illustrates their potential of becoming the possible drug targets. Therefore PSORTb tools at Expasy server was utilized to identify the subcellular localization of non-homologous essential protein sequences (Yu et al., 2010). The modulation of the activity of a protein target with a small molecule of a drug accounts for its prospective druggability. Drug Bank Database was accessed to calculate the druggability potential of each identified drug target (Knox et al., 2011). BLASTp with default parameters was used to align the potential drug targets from Ralstonia solanacearum against the list of the of compounds found within the Drug Bank (Szklarczyk et al., 2019).
Selected indispensable proteins were then subjected to STRING database (http://string.embl.de) to construct protein-protein interaction network (Li, Jaroszewski and Godzik, 2001). Interactors with confidence score greater than or equal to 0.700 alone included here in the protein network and with low and medium confidence score were eliminated to avoid false positive and false negative results. Target protein with more interactors is considered as a metabolically active protein which could serve as appropriate Drug target (Peyraud et al., 2017; Szklarczyk et al., 2019).
RESULTS AND DISCUSSION
The main goal of the subtractive genomic analysis was to examine Rolastonia solanacearum GM1000 strain critical proteins as a possible drug target for future strategic drug discovery. Total 5106 proteins of total proteome were originally obtained from Ralstonia solanacearum GM1000 Uniprot database. The CD-HIT tool was used to differentiate paralogous and non-paralogous proteins. 134 paralogous proteins were screened and 4972 non paralogous sequence of proteins were selected for further analysis.
The selected proteins were assessed against Solanaceae proteome in BLASTp, with an E-value cut off 10-3. Selected non homologous proteins were employed for the identification of essential gene using BLASTp tools of DEG database at default parameter settings. The analysis identified 465 essential proteins. There are 41 hypothetical proteins were identified which were finally excluded in this study. The essential proteins of bacteria are expected to be involved in housekeeping and are important for the survival of pathogen.
Total no of protein 5106
Duplicate (>80% identical) in CD-HIT 134
Essential proteins in DEG (E-value 10-5) 465
Number of hypothetical proteins as essential proteins 41
Essential proteins involved in metabolic pathway 117
Unique metabolic pathway essential proteins 7
Essential proteins found to be druggable 4
Functional pathway assessment of 424 essential proteins were conducted using KAAS server. Among 424 proteins 117 proteins were found to be involved in different metabolic pathway of the pathogen. These 117 proteins were further analyzed by the BLASTp algorithm for the comparison of metabolic pathway in Ralstonia solanacearum proteome and Solanum tuberosum proteome as a reference organism of Solanaceae family to exclude the common pathway. Total seven pathogen specific pathways of Ralstonia solanacearum GM 1000 were identified by KEGG which were absent in Solanaceae family. Total seven nonhomologous proteins were identified.
that are thought to be essential and involved in pathogens unique metabolic pathway. Therefore, new drugs may be designed to target these essential proteins to inhibit one or more of these metabolic pathways thereby controlling the growth and viability of the pathogenic strain Ralstonia solanacearum GM 1000. The total seven non homolog essential proteins (Table1) so obtained were verified within Drug Bank Database for possible druggability and four essential non homologous proteins (Table 2) were identified to have druggability potential. Thereafter, the four selected proteins were then subjected to PSORTb for their sub cellular localization.
Table 1. Unique metabolic pathway essential proteins
| Sl no | DEG ID | UNIPROT ID/ DRUGGABILITY | METABOLIC PATHWAY |
| 1 | DEG10570448 | Q8XW91
Druggable |
QUORUM SENSING
|
| 2. | DEG10570275 | Q8XX10
Druggable |
BACTERIAL SECRETION SYSTEM |
| 3. |
DEG10570247 |
Q8Y3B8
Druggable |
PEPTITOGLYCAN BIOSYNTHESIS
BETA LACTAM RESISTANCE |
| 4. |
DEG10570255 |
Q8XVI1
Druggable |
BETA LACTAM RESISTANCE
PEPTIDOGLYCAN BIOSYNTHESIS |
| 5. |
DEG10570232 |
Q8XQ85
Not Druggable |
BACTERIAL CHEMOTAXIS |
| 6. |
DEG10570446 |
Q8XVG2
Not Druggable |
QUORUM SENSING |
| 7. |
DEG10570220 |
Q8XX15
Not Druggable |
BACTERIAL SECRETION SYSTEM |
Sub Cellular Localization
| Name of Protein | Uniprot ID | Location |
| Probable conjugal transfer protein trbb | Q8XW91 | cytoplasmic |
| Probable type II secretory pathway gspe-related protein (RSc2308) | Q8XX10 | cytoplasmic |
| Peptidoglycan D, D-transpeptidase MrdA | Q8Y3B8 | cytoplasmic |
| Peptidoglycan D, D-transpeptidase FtsI | Q8XVI1 | Cytoplasmic Membrane |
Table 2. Non homologous essential protein of Ralstonia solanacearum strain similar to binding pattern of FDA approved drugs against DrugBank database using BLASTp
| Sl. no | Protein name | DrugBank ID | Uniprot ID |
| 1. | Conjugal transfer protein trbb | DB02930
DB04395 |
Q8XW91 |
| 2. | Type II secretory pathway gspe-related protein (RSc2308) | DB04395
DB02930 |
Q8XX10 |
| 3. | Peptidoglycan D, D-transpeptidase MrdA | DB01413, DB00438, DB14879, DB01598, DB01329, DB01327, DB01163, DB01163, DB01328, DB01413, DB01415, DB00948, DB00438, DB00303, DB00671, DB01326, DB00923DB00355, DB00493, DB04570, DB01413, DB01147, DB09050, DB06211, DB14879
DB04918DB00274, DB00430, DB01607, DB01000 DB02443, DB02968, DB04041, DB01603, DB00417
|
Q8Y3B8 |
| 4. | Peptidoglycan D, D-transpeptidase FtsI
|
DB01413, DB01147, DB09050, DB06211, DB14879DB04918, DB00267, DB01416, DB01329, DB01327, DB01331, DB01328, DB01413, DB01415, DB00430DB05659DB00535, DB04918, DB01150DB03190 |
Q8XVI1 |
Earlier, 20 proteins of Ralstonia solanacearum were targeted for drug design having Protein Data Bank (PDB) ID of 3ZI8, 4I68, 4KF9, 4FDB, 3UMB, 3TMB, 3TOT, 3TOU, 3NPN, 3NPQ, 3LOP, 3GG9, 3GHY, 3EN2, 2QGU, 2CHH, 2BT9, 2BS5, 2BS6, 1UQX. (Kotaki and Saikia, 2015). Peptidoglycan D, D-transpeptidase MrdA, Peptidoglycan D, D-transpeptidase FtsI, Type II secretory pathway gspe-related protein were identified as the best predicted protein for drug target in this study. Type II secretion system is a virulent factor of R. solanacearum (Peeters et al., 2013). Inhibition of Quarum sensing protein can only prevent biofilm formation of pathogenic bacteria without any apparent direct effect on survivability. However, Peptidoglycan D, D-transpeptidase MrdA,
Peptidoglycan D, D-transpeptidase FtsI protein as drug targets have already been reported and efforts have been taken for drug design in many human pathogenic bacterial strains, but these drug targets are inapplicable for Ralstonia solanacearum strains as β lactam antibiotics are less effective in controlling bacterial wilt disease (Souvage and Terrak, 2016; Waack et al., 2017). Different Secretion systems of bacteria are very attractive targets for alternative therapeutics because their inactivation interferes with the delivery of secreted virulence factors. There are many cell walls degrading enzymes are secreted through Type II secretory system (T2SS) in Ralstonia solanacearum. Therefore, inhibitor of Type II secretory system (T2SS) could be a good alternative for drug design.
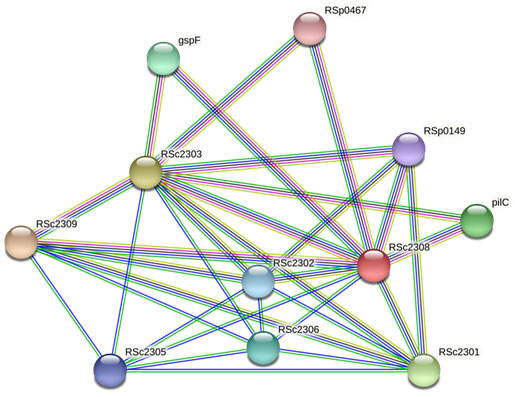
Rsc2308 (UniProtKB ID- Q8XX10) is the Type II secretory pathway gspe-related protein of Ralstonia solanacearum associated with secretory system of bacteria which is responsible for pathogenicity. Therefore. Type II secretory pathway gspe-related protein (RSc2308) of Ralstonia solanacearum could be a promising drug target for future drug design that has not been properly addressed so far. Network based analysis showed that this protein Rsc3208 is interconnected with eighteen proteins in network with combined score greater than 0.7 (Table3) (Salanoubat et al., 2001; Waack et al., 2017).
So, it may be assumed that this Type II secretory pathway gspe- related protein is a highly metabolically active protein and inhibition of this protein may arrest the growth of the bacteria. Therefore, the present work opens a new avenue for searching novel drug compounds that may interact with the target Type II secretory pathway gspe-related protein (RSc2308) and may pave the path for new control strategy (Souvage and Terrak, 2016).
Figure 1
Figure 1: Interaction among Type II secretory pathway gspe-related protein (RSc2308) and other proteins of R. solanacearum.
Table 3. Interaction among Type II secretory pathway gspe-related protein (RSc2308) and other proteins of R. solanacearum and their combined score.
| node1 | node2 | node1 string id | node2 string id | combined_score |
| RSc2300 | RSc2308 | 267608.RSc2300 | 267608.RSc2308 | 0.762 |
| RSc2301 | RSc2308 | 267608.RSc2301 | 267608.RSc2308 | 0.922 |
| RSc2302 | RSc2308 | 267608.RSc2302 | 267608.RSc2308 | 0.886 |
| RSc2303 | RSc2308 | 267608.RSc2303 | 267608.RSc2308 | 0.955 |
| RSc2304 | RSc2308 | 267608.RSc2304 | 267608.RSc2308 | 0.867 |
| RSc2305 | RSc2308 | 267608.RSc2305 | 267608.RSc2308 | 0.884 |
| RSc2306 | RSc2308 | 267608.RSc2306 | 267608.RSc2308 | 0.887 |
| RSc2307 | RSc2308 | 267608.RSc2307 | 267608.RSc2308 | 0.845 |
| RSc2309 | RSc2308 | 267608.RSc2309 | 267608.RSc2308 | 0.981 |
| RSc2310 | RSc2308 | 267608.RSc2310 | 267608.RSc2308 | 0.869 |
| RSp0143 | RSc2308 | 267608.RSp0143 | 267608.RSc2308 | 0.772 |
| RSp0149 | RSc2308 | 267608.RSp0149 | 267608.RSc2308 | 0.884 |
| RSp0467 | RSc2308 | 267608.RSp0467 | 267608.RSc2308 | 0.882 |
| RSp0474 | RSc2308 | 267608.RSp0474 | 267608.RSc2308 | 0.715 |
| gspD | RSc2308 | 267608.RSc3114 | 267608.RSc2308 | 0.756 |
| gspF | RSc2308 | 267608.RSc3116 | 267608.RSc2308 | 0.895 |
| pilC | RSc2308 | 267608.RSc2826 | 267608.RSc2308 | 0.896 |
| pilD | RSc2308 | 267608.RSc2827 | 267608.RSc2308 | 0.790 |
CONCLUSION
Subtractive genome analysis revealed possible drug targets in many human pathogenic bacteria and only few in plant pathogenic bacteria. In silico identification of possible drug target in Ralstonia solanacearum is completely lacking. Therefore, the present work probably is the first report underpinning the druggability of type II secretory pathway gspe-related protein of Ralstonia solanacearum through subtractive genome analysis. The gspe-related protein is essential in type-2 secretion pathway for secreting cell wall degrading enzymes that are key to host penetration and colonization. Therefore, targeting the protein with new drugs may prevent host colonization and survival in the weeds thereby offering a good strategy for controlling the pathogen in future.
ACKNOWLEDGEMENTS
The present work has not been supported financially by any funding agencies. The authors would like to acknowledge Department of Botany, Vivekananda Mahavidyalaya, Haripal Hooghly for necessary support.
Conflict of Interest:The authors declare that there is no conflict of interests.
REFERENCES
Allen C, Bent A, and Charkowski A., (2009). Underexplored niches in research on plant pathogenic bacteria. Plant Physiol., 150(4), 1631–1637.
Amineni, U., Pradhan, D. and Marisetty, H., (2010). In silico identification of common putative drug targets in Leptospira interrogans. J Chem Biol, 3(4), 165–173.
Arndt, D., Wilson, M., Neveu, V., Tang, A., Gabriel, G., Ly, C., Adamjee, S., Dame, Z. T., Han, B., Zhou, Y. and Wishart, D. S., (2014). DrugBank 4.0: shedding new light on drug metabolism. Nucleic acids research, 42, D1091-D1097.
Barh D., Tiwari S., Jain N., Ali A., Santos A.R., Misra A.N, Azevedo V. and Kumar A., (2011). In silico subtractive genomics for target identification in human bacterial pathogens. Drug Dev Res. 7, 162–177.
Coutinho, T. A. and Wingfield, M. J. (2017). Ralstonia solanacearum and R. pseudosolanacearum on Eucalyptus: Opportunists or Primary Pathogens? Frontiers in Plant Science, 8, 761.
Elphinstone JG., (2005). The current bacterial wilt situation: a global overview. In: Allen C, Prior P, Hayward AC, (2005). Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex. American Phytopathological Society Press; St Paul, MN, 9–28.
Gadewar, A. V., Trivedi T. P. and Sekhawat G. S., (1991). Potato in Karnataka. Tech. Bull, 17, 33.
Ishikawa, R., Shirouzu, K., Nakashita, H., Teraoka, T., and Arie, T., (2007). Control efficacy of validamycin A against Fusarium wilt correlated with the severity of phytotoxic necrosis formed on tomato tissues. Journal of Pesticide Science, Volume 32, Issue 2, Pages 83-88.
Kataki, M., and Saikia M. K., (2015). In silico binding studies of oxalis corniculata compounds with Ralstonia solanacearum proteins and histone deacetylase 8 protein. International Journal of Drug Research and Technology, Vol. 5 (1), 13-23.
Keshri V., Singh DP, Prabha R., Rai A. and Sharma AK, (2014). Genome subtraction for the identification of potential antimicrobial targets in Xanthomonas oryzae pv. oryzae PXO99A pathogenic to rice. 3 Biotech., 4(1), 91–95.
Khanum SA, Shashikanth S., Umesha S., and Kavitha R., (2005). Synthesis and antimicrobial study of novel heterocyclic compounds from hydroxybenzophenones. European Journal of Medicinal Chemistry. 40(11), 1156-1162.
Law, V., Knox, C., Djoumbou, Y., Jewison, T., Guo, A. C., Liu, Y., Maciejewski, A., Arndt, D., Wilson, M., Neveu, V., Tang, A., Gabriel, G., Ly, C., Adamjee, S., Dame, Z. T., Han, B., Zhou, Y. and Wishart, D. S., (2014). DrugBank 4.0: shedding new light on drug metabolism. Nucleic acids research, 42, D1091-D1097.
Lee YH, Choi CW, Kim SH, Yun JG, Chang SW, Kim YS, and Hong JK, (2012). Chemical pesticides and plant essential oils for disease control of tomato bacterial wilt. Plant Pathol J., 28, 32–39.
Li, W., Jaroszewski, L. and Godzik, A., (2001). Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics, 17, 282-283.
Lin Y, He Z, Rosskopf EN, Conn KL, Powell CA and Lazarovits G., (2010). A nylon membrane bag assay for determination of the effect of chemicals on soil borne plant pathogens in soil. Plan Dis. 94, 201–206.
Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I., Salmond G., and Foster GD, (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol, 13(6) 614-29.
Mbaka, J. N., J. K. Gitonga, C. W. Gathambari, B. G. Mwangi, P. Githuka and M. Mwangi., (2013). Identification of knowledge and technology gaps in high tunnels tomato production in Kirinyaga and Embu counties.
Miesel L., Greene J., and Black TA, (2003). Genetic strategies for antibacterial drug discovery. Nat Rev Genet, 4(6), 442–456.
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C. and Kanehisa, M., (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res, 35, W182-5.
Peeters, N., Guidot, A., Vailleau, F. and Valls, M., (2013). Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol Plant Pathol, 14, 651-62.
Peyraud, R., Dubiella, U., Barbacci, A., Genin, S., Raffaele, S. and Roby, D., (2017). Advances on plant–pathogen interactions from molecular toward systems biology perspectives. The Plant Journal, 90(4), pp.720-737.
Prabha, R., Singh, D.P., Ahmad, K. Kumar, S.P.J. and Kumar, P., (2019). Subtractive genomics approach for identification of putative antimicrobial targets in Xanthomonas oryzae pv. oryzae KACC10331. Arch. Phytopath. Plant Protect. 52, 863–872.
Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, Billault A, Brottier P, Camus JC, Cattolico L, Chandler M, Choisne N, Claudel-Renard C, Cunnac S, Demange N, Gaspin C, Lavie M, Moisan A, Robert C, Saurin W, Schiex T, Siguier P, Thébault P, Whalen M, Wincker P, Levy M, Weissenbach J. and Boucher CA, (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature, 31;415(6871), 497-502.
Santana M., (2020). In Silico Approaches for Prioritizing Drug Targets in Pathogens. In: Panwar H., Sharma C., and Lichtfouse E. (2020) Sustainable Agriculture Reviews 46. Sustainable Agriculture Reviews, vol 46, Springer, Cham.
Sauvage E, and Terrak M., (2016). Glycosyltransferases and Transpeptidases/Penicillin-Binding Proteins: Valuable Targets for New Antibacterials. Antibiotics (Basel), 17; 5(1), 12.
Schell MA., (2000). Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory array. Ann Rev Phytopathol, 38, 263–292.
Silver LL., (2011). Challenges of antibacterial discovery. Clin Microbiol Rev., 24(1), 71–109.
Somani, A. K., Chakrabarti, S. K., and Pandey and S. K., (2010). Spread of bacterial wilt and brown rot of potato in Indore region of Madhya Pradesh. CPRI News Letter no., 42 (June), 16–17.
Sudha, R., Katiyar, A., Katiyar, P., Singh, H., and Prasad, P., (2019). Identification of potential drug targets and vaccine candidates in Clostridium botulinum using subtractive genomics approach. Bioinformation, 15(1), 18–25.
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ and Mering CV., (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res., 8; 47(D1): D607-D613.
The Uniprot, C., (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research, 47, D506-D515.
Van Elsas, J. D., P. Kastelein, P. M. de Vries and L. S. van Overbeek. (2001). Effects of ecological factors on the survival and physiology of Ralstonia solanacearum bv. 2 in irrigation water. Can. J. Microbiol. 47, 842-854.
Vetrivel U., Subramanian G. and Dorairaj S., (2011). A novel in silico approach to identify potential therapeutic targets in human bacterial pathogens. Hugo J., 5(1–4), 25–34.
Waack U, Johnson TL, Chedid K, Xi C, Simmons LA, Mobley HLT and Sandkvist M., (2017). Targeting the Type II Secretion System: Development, Optimization, and Validation of a High-Throughput Screen for the Identification of Small Molecule Inhibitors. Front Cell Infect Microbiol, 28;7, 380.
Wang JF, and Lin CH., (2005). Integrated management of tomato bacterial wilt. AVRDC-The World Vegetable Center, Taiwan.
Whipps JM, and Gerhardson B., (2007). Biological pesticides for control of seed- and soil-borne plant pathogens. A training course guide. In: Van Elsas JD, Jansson JD, Trevors JT (eds) Modern soil microbiology, 2nd edn. CRC Press, Boca Raton, 479–501.
Wicker, E., Grassart, L., Coranson-Beaudu, R., Mian, D., Guilbaud, C. and Fegan, M., Prior P., (2007). Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Appl Environ Microbiol, 71, 6790–6801.
Yan F, and Gao F., (2020). A systematic strategy for the investigation of vaccines and drugs targeting bacteria. Comput Struct Biotechnol J., Jun 12; 18, 1525-1538.
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, and Brinkman FS., (2010). PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics, 1;26(13), 1608-15.
Yuliar, Nion, Y. A. and Toyota, K., (2015). Recent Trends in Control Methods for Bacterial Wilt Diseases Caused by Ralstonia solanacearum. Microbes and Environments, 30, 1-11.