Department of Biological Science, Faculty of Science, King Abdulaziz
University, Jeddah, Kingdom of Saudi Arabia.
Corresponding author email: ahmadfirozbin@gmail.com
Article Publishing History
Received: 25/09/2021
Accepted After Revision: 24/12/2021
Proliferative diabetic retinopathy is the widespread type of DM which causes chronic as well as progressive alterations at microvascular level, which particularly effects the eye. The main characteristic of this disease is the development of few new blood vessels around the retina of eye as well as at the posterior region of eye segments. For our computational analysis 155 differentially expressed genes calculated through paired t test statistics analysis using the GenePattern platform, of proliferative diabetic retinopathy in Saudi patients were downloaded. Among the 155 genes, 95 were upregulated, and 60 were downregulated. The Annotation Cluster (FAC) tool in the (DAVID)
(http://david.abcc.ncifcrf.gov/home.jsp) was used to identify biological processes that are abundant in proliferative diabetic retinopathy (PDR). The functions required for response to mRNA splicing, intracellular protein transport, mRNA processing, microtubule cytoskeleton structure, and atrioventricular canal formation are represented by the GO keywords that are abundant in genes. We used the KAAS web server to identify the biological pathways of these DEGs in addition to DAVID functional analysis and found that the majority of the DEGs were associated with important biological processes, with many being classified in metabolic pathways, Spliceosome, Cell cycle, or being involved in the mRNA surveillance pathway. findings are consistent with those of earlier research. To corroborate the predictions stated in this work, which will demonstrate the role enhanced functional processes, experimental validation will be necessary.
Computational Analysis, Diabetes Mellitus, Gene Expression, Proliferative Diabetic Retinopathy
Alzahrani A. B, Ali H. M, Firoz A. Gene Expression in Proliferative Diabetic Retinopathy using RNA-Seq Data: A Computational Study on Saudi Patients. Biosc.Biotech.Res.Comm. 2021;14(4).
Alzahrani A.B, Ali H.M, Firoz A. Gene Expression in Proliferative Diabetic Retinopathy using RNA-Seq Data: A Computational Study on Saudi Patients. Biosc.Biotech.Res.Comm. 2021;14(4). Available from: <a href=”https://bit.ly/31bFKFa“>https://bit.ly/31bFKFa</a>
Copyright © Alzahrani and Firoz This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Diabetes mellitus is one of the multifactorial diseases and a leading cause of death in world and especially in Saudi Arabia. Proliferative diabetic retinopathy is the widespread type of DM which causes chronic as well as progressive alterations at microvascular level, which particularly effects the eye, along with other body parts. If the disease is left untreated it will grow gradually and ultimately leading to the blindness. Progression of disease is not rapid, but gradual starting from mild alterations, moving towards moderate and ultimately severe proliferative diabetic retinopathy. The main characteristic of this disease is the development of few new blood vessels around the retina of eye as well as at the posterior region of eye segments i.e., vitreous (El-Bab et al. 2012; Lee et al. 2015; Alharbi and Alhazmi 2020).
The mechanism by which the DM progresses to diabetic retinopathy is not clearly understood and that’s why the disease pathology is thought to be complex and unclear. However, a lot of studies has been carried out to examine the disease progression by considering the disease history along with other aspects. It has been suggested that multiple interactive mechanisms are playing an important role, causing the damage at cellular level and adaptive changes, which cause the devastation in this disease(El-Asrar et al. 1998; Sinclair and Schwartz 2019; Alharbi and Alhazmi 2020).
Earlier it was considered that DM and especially PDR is not a prevalent disease at Saudi Arabia, due to healthy diet and routine. However, recent studies have reported that prevalence of disease is increasing in Saudi Arabia as bell and the possible risk factors for this progression are supposed to be consumption of more westernized diet leading to increased chances of obesity and ultimately complications of diabetes. Earlier the disease was 23.7% prevalent in Saudi Arabia while by the year (2011), it has reached to increase 30% and increasing day by day with men more affected that females (Ali et al. 2008; Al Dawish et al. 2016; Alharbi and Alhazmi 2020).
Different treatment strategies can be used to treat diabetic retinopathy. Photocoagulation is one of them. Studies has shown that photocoagulation approach causes a decrease in chances of loss of vision by up to 50% (Cantrill 1984). It causes the decrease in visual acuity as well as constricts the posterior visual regions. Intravitreal administration of about 1.25 mg bevacizumab at the time of cataract surgery could be safe as well as protective in preventing the progression of DR and diabetic maculopathy in patients with cataract and diabetic retinopathy (Cheema et al. 2009; Alghamdi et al. 2021).
MATERIAL AND METHODS
155 differentially expressed genes calculated through paired t test statistics analysis using the GenePattern platform, and identified based on the statistical cutoff of proliferative diabetic retinopathy in Saudi patients with type 2 diabetes were downloaded (Pan et al. 2016). Among the 155 genes, 95 were upregulated, and 60 were downregulated, and has been taken for computational analysis shown in Table 1.
For the functional analysis, on the list of differentially expressed genes with a fold change of >1, DAVID (http://david.abcc.ncifcrf.gov/home.jsp) functional annotation cluster analysis was done. For analysis, only terms with a value of 0.05 and a count number of 5 genes were chosen. DAVID was used to classify enriched biological themes in the collection of DEGGs using the gene ontology (GO) term biological process (BP). The KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/kegg/kaas/) was used to map pathways (Moriya et al. 2007).
The amino acid sequences of these DEGGs were submitted to the KAAS online site as input, and orthologs were assigned using the single-directional best hit (SBH) technique. KAAS uses BLAST similarity searches against a carefully selected set of ortholog groups in the KEGG GENES database to provide functional annotation of genes in a genome. Genes in the data sets that were mapped to one of KEGG’s reference pathways were given a KEGG orthology (KO) number by KAAS (Amoaku et al. 2020).
Table 1. A list of 155 differentially expressed gene selected for analysis (Pan et al. 2016).
| ZNF207 | UTRN | CTR9 | TRPS1 | ZNF80 | CAV2 | IL33 |
| SMAD4 | TARDBP | YTHDC1 | USP8 | KCNH3 | LOC285847 | ADRA1D |
| SEPT2 | SFPQ | MIA3 | MEAF6 | LOC100506124 | RAMP1 | TAAR1 |
| ORC2 | CEP350 | UBL4A | FAM69B | RNPC3 | LOC100506995 | OR4K17 |
| MTCH1 | FAM208B | GPR18 | MGC72080 | TUBG1 | PRSS27 | KDR |
| SENP1 | ECD | KCTD4 | RBM17 | PSAT1 | GPRC5D | LOC440910 |
| HELZ | CORO7 | BTBD2 | RQCD1 | PCDHGC4 | NCKAP5 | |
| TTC17 | EI24 | TUG1 | MMGT1 | DLC1 | HLA-DQA2 | |
| MAN2A1 | STARD3 | C11orf30 | LATS1 | DDX11L9 | LINC00346 | |
| KLHL11 | SYTL1 | ZNF597 | WEE1 | CTHRC1 | RCVRN | |
| PTPN11 | EIF3A | SEC63 | HIST1H3I | RNU4-2 | DYX1C1 | |
| SSR1 | ASTE1 | STX17 | SEPN1 | DPY19L2 | MFAP5 | |
| DUSP11 | CHMP6 | VPS36 | CLOCK | NTSR1 | OVCH1 | |
| TMED10 | HNRNPA3 | PSMD5 | LINC00265 | IFITM10 | PNMT | |
| DYNC1LI2 | PSIMCT-1 | FAM190B | PPP6C | ACY3 | PPP1R14C | |
| SLC39A3 | TMEM39A | DTWD2 | LOC729852 | FP588 | CCDC144NL | |
| SLC33A1 | CCND3 | RBM25 | PAICS | NXPH3 | LOC100505806 | |
| ARHGEF6 | PACS2 | PRIM2 | LOC100652890 | LOC100506476 | GS52 | |
| PAPOLG | PDE4D | ZNF784 | HSP90B1 | NRXN2 | HRH3 | |
| HNRNPU | KIAA2026 | CG030 | EYS | LOC100506678 | PP2672 | |
| ASH1L | CALU | ABRA | MOB3B | LOC100507144 | CPSF4L | |
| LOC729082 | AP3M1 | PDCD4 | PNN | PNMAL2 | RTL1 | |
| USP48 | MASP2 | CCAR1 | HNRNPA2B1 | BCL6B | LOC100128081 | |
| ZDHHC6 | ALG2 | SOS1-IT1 | SDHAP2 | TDRD10 | CNPY1 | |
| UTRN | DDX46 | LOC283624 | RAB40B | HCAR1 | UGT2A3 |
RESULTS AND DISCUSSION
We downloaded the precomputed list of 155 differentially expressed genes for our computational analysis shown in Table-1Among the 155 genes, 95 were upregulated, and 60 were downregulated (Pan et al. 2016; Amoaku et al. 2020).
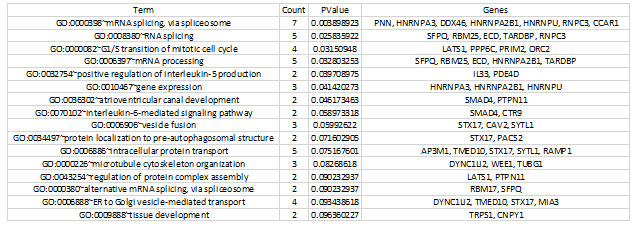
For the functional annotation analysis, the Annotation Cluster (FAC) tool in the Database for Annotation, Visualization, and Integrated Discovery (DAVID) was used to identify biological processes that are enriched in proliferative diabetic retinopathy (PDR) (http://david.abcc.ncifcrf.gov/home.jsp). For annotations and GO terms with statistically significant values from the resultant functional analysis, the name “Biological Process” was utilized. The functions required for response to mRNA splicing, intracellular protein transport, mRNA processing, microtubule cytoskeleton structure, and atrioventricular canal formation are represented by the GO keywords that are abundant in genes in this table (Table 2).
Table 2. Significantly enriched gene ontology (GO) terms detected by FAC in differentially expressed genes.
Only those terms which reported a value of ≤0.05 and count number ≥2 genes were selected for the analysis.
For the pathway analysis, we found the biological pathways of DEGs annotated in the current study in addition to DAVID functional analysis. DEG amino acid sequences in FASTA format were put into the KAAS to predict different pathways. There was a total of 154 routes predicted. Table 3 lists the top 20 KEGG pathways, with Supplementary Table S1 providing a comprehensive list of all pathways. The majority of DEGs were discovered to relate to significant biological processes, with many being categorized in metabolic pathways, spliceosomes, or cell cycle, or being engaged in the mRNA monitoring pathway, as seen in these tables (Amoaku et al. 2020).
Table 3. Top 20 KEGG pathways for DEGs, Number of mapped genes shown in bracket
| ko01100 Metabolic pathways (10) |
| ko03040 Spliceosome (5) |
| ko04110 Cell cycle (4) |
| ko05164 Influenza A (4) |
| ko04144 Endocytosis (4) |
| ko01110 Biosynthesis of secondary metabolites (4) |
| ko05205 Proteoglycans in cancer (4) |
| ko04080 Neuroactive ligand-receptor interaction (4) |
| ko03015 mRNA surveillance pathway (4) |
| ko05132 Salmonella infection (3) |
| ko05200 Pathways in cancer (3) |
| ko04141 Protein processing in endoplasmic reticulum (3) |
| ko04151 PI3K-Akt signaling pathway (3) |
| ko05166 Human T-cell leukemia virus 1 infection (3) |
| ko04020 Calcium signaling pathway (3) |
| ko05168 Herpes simplex virus 1 infection (3) |
| ko05207 Chemical carcinogenesis – receptor activation (3) |
| ko04510 Focal adhesion (3) |
| ko04390 Hippo signaling pathway (3) |
| ko05418 Fluid shear stress and atherosclerosis (3) |
| ko05014 Amyotrophic lateral sclerosis (3) |
Recent studies have reported prevalence of Proliferative diabetic retinopathy (PDR) disease is increasing. Proliferative diabetic retinopathy is the widespread type of DM which causes chronic as well as progressive alterations at microvascular level, which particularly effects the eye. The main characteristic of this disease is the abnormal growth of new vessels occurs (Tarr et al. 2013; Safi et al. 2014). Study shows Involvement of angiogenesis, inflammation, and fibrosis in proliferative diabetic retinopathy and Enrichment of genes and pathways related to lymphatic development indicates that targeting lymphatic involvement in PDR progression.
Several pro-angiogenic cytokines have been described as being involved in the pathogenesis of PDR, although VEGF is accepted as the most significant cytokine in PDR (Amoaku et al. 2020). The present finding shows significance of mRNA splicing, intracellular protein transport, mRNA processing, microtubule cytoskeleton organization and atrioventricular canal development, and associated with important biological processes, many being classified in metabolic pathways, Spliceosome, Cell cycle or being involved in mRNA surveillance pathway These are consistent with those of other studies (Korhonen et al. 2021).
CONCLUSION
The findings of the present study have used a Bioinformatics approach to identify the DEGs enrichment indicate the significance of mRNA splicing, intracellular protein transport, mRNA processing, microtubule cytoskeleton organization and atrioventricular canal development, and associated with important biological processes, many being classified in metabolic pathways, Spliceosome, Cell cycle or being involved in mRNA surveillance pathway The present study’s findings are consistent with those of earlier research. To corroborate the predictions stated in this work, which will demonstrate the role enhanced functional processes, experimental validation will be necessary.
ACKNOWLEDGEMENTS
This study was technologically supported by the Bioinformatics and Computational Biology Unit at Department of Biological Sciences, King Abdulaziz University, Jeddah, Saudi Arabia.
REFERENCES
AL DAWISH, M. A., ROBERT, A. A., BRAHAM, R. et al. (2016). Diabetes Mellitus in Saudi Arabia: A Review of the Recent Literature. Curr Diabetes Rev, 12, 359-368.
ALGHAMDI, S. A., TOURKMANI, A. M., ALHARBI, T. J et al. (2021). Prevalence of retinopathy and associated risk factors among high- and low-risk patients with type 2 diabetes mellitus. An observational study, 42, 693-697.
ALHARBI, A. M. D. and ALHAZMI, A. M. S. (2020). Prevalence, Risk Factors, and Patient Awareness of Diabetic Retinopathy in Saudi Arabia: A Review of the Literature. Cureus, 12, e11991.
ALI, M., WHITE, J., LEE, C. H., et al. (2008). Therapy conversion to biphasic insulin aspart 30 improves long-term outcomes and reduces the costs of type 2 diabetes in Saudi Arabia. J Med Econ, 11, 651-70.
AMOAKU, W. M., GHANCHI, F., BAILEY, C et al. (2020). Diabetic retinopathy and diabetic macular oedema pathways and management: UK Consensus Working Group. Eye, 34, 1-51.
CANTRILL, H. L. (1984). The diabetic retinopathy study and the early treatment diabetic retinopathy study. Int Ophthalmol Clin, 24, 13-29.
CHEEMA, R. A., AL-MUBARAK, M. M., AMIN, Y. M. et al. (2009). Role of combined cataract surgery and intravitreal bevacizumab injection in preventing progression of diabetic retinopathy: prospective randomized study. J Cataract Refract Surg, 35, 18-25.
EL-ASRAR, A. M., AL-RUBEAAN, K. A., AL-AMRO, S. A.(1998). Risk factors for diabetic retinopathy among Saudi diabetics. Int Ophthalmol, 22, 155-61.
EL-BAB, M. F., SHAWKY, N., AL-SISI, A. (2012). Retinopathy and risk factors in diabetic patients from Al-Madinah Al-Munawarah in the Kingdom of Saudi Arabia. Clin Ophthalmol, 6, 269-76.
KORHONEN, A., GUCCIARDO, E., LEHTI, K. et al. (2021). Proliferative diabetic retinopathy transcriptomes reveal angiogenesis, anti-angiogenic therapy escape mechanisms, fibrosis and lymphatic involvement. Scientific Reports, 11, 18810.
LEE, R., WONG, T. Y. and SABANAYAGAM, C. (2015). Epidemiology of diabetic retinopathy, diabetic macular edema and related vision loss. Eye and Vision, 2, 17.
PAN, J., LIU, S., FARKAS, M et al. (2016). Serum molecular signature for proliferative diabetic retinopathy in Saudi patients with type 2 diabetes. Mol Vis, 22, 636-45.
SAFI, S. Z., QVIST, R., KUMAR, S., (2014). Molecular mechanisms of diabetic retinopathy, general preventive strategies, and novel therapeutic targets. BioMed research international, 2014, 801269-801269.
SINCLAIR, S. H. and SCHWARTZ, S. S. (2019). Diabetic Retinopathy–An Underdiagnosed and Undertreated Inflammatory, Neuro-Vascular Complication of Diabetes. Frontiers in Endocrinology, 10.
TARR, J. M., KAUL, K., CHOPRA, M et al. (2013). Pathophysiology of Diabetic Retinopathy. ISRN Ophthalmology, 2013, 343560.


