Postgraduate Department of Botany and Research Centre, University College, Thiruvananthapuram-695034, India
Corresponding author email: subr44@gmail.com
Article Publishing History
Received: 27/10/2020
Accepted After Revision: 14/12/2020
Microorganisms and their enzymes are important part of industrial biotechnology because of their high adaptability, versatile metabolic machinery and simple genetic constitution which can be easily manipulated to meet different needs. A mesophilic novel strain of Serratia marcescens – VT1 was found to produce extracellular protease (13.173 U/ml) and cold active and stable psychrophilic lipase (23.17 U/ml). Characterization of protease showed the enzyme to be active over a wide range of pH 4-11, optimum pH 10, and optimum temperature 50°C. SMVT1 lipase was found to be active from pH 7-9 with optimum pH 7 and good stability for 90 minutes, optimum temperature 30°C and to retain almost 80 % activity at lower temperatures (20 and 10°C). SMVT 1 protease was stable in hydrophilic solvents like methanol and ethanol. Lipase showed stability in both hydrophobic and hydrophilic solvents, hexane- 92.67%, methanol-87.4% and acetone-85.23%. The wide range pH active protease and psychrophilic and organic solvent stable lipase can be made use in various sectors like bioremediation, waste management, and chemical synthesis, food processing and detergent additives.
Serratia marcescens, Protease, Lipase, Psychrophilic, Solvent Stability.
Vivek K, Sandhia G. S, Subramaniyan S. Screening, Production and Characterization of Industrially Important Enzymes by Serratia marcescens Strain VT 1. Biosc.Biotech.Res.Comm. 2020;13(4).
Vivek K, Sandhia G. S, Subramaniyan S. Screening, Production and Characterization of Industrially Important Enzymes by Serratia marcescens Strain VT 1. Biosc.Biotech.Res.Comm. 2020;13(4). Available from: <a href=”https://bit.ly/3mGFfrw”>https://bit.ly/3mGFfrw</a>
Copyright © Vivek et al., This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
The importance of microbial enzyme lies in the role played by these bio-molecules in catalyzing the broad array of reactions which is indispensable for maintaining life in earth. Enzymes can be isolated from any living forms and each one has its own characteristic property which separates it from other. In industrial sector enzymes have immense applications, among these enzymes microbial ones form an inevitable part and have always been in demand due to their unique properties (Liu and Kokare 2017). There is an increase the likelihood for finding enzymes with distinctive characters in area ranging from dry deserts to extreme cold Antarctic regions. Microbial enzymes are known to be more stable and catalyse a wide range of activities when compared to their counterparts (Hasan et al., 2006). Microbial enzymes have applications in food and beverage, diary, leather, paper and pulp, pharmaceuticals, fertilizer and detergent industries. They are also used for degradation and sustainable management of waste materials and production of bio-fuels. Among these enzymes protease remains the dominant one till to date followed by cellulase and lipase, (Chapman et al., 2018 Eddehech et al. 2019 Chandra et al 2020).
Proteases (EC 3.4.21) are enzyme that increases the rate of proteolysis resulting in single amino acids, the end product of protein breakdown resulting from the cleavage of peptide bond (Barrett and McDonald, 1986). Around 60 % of enzyme that is being marketed worldwide is protease (Rao et al., 2009). About two third of the commercial protease is of microbial origin (Beg and Gupta 2003). Lipases (EC 3.1.1.3) are hydrolytic enzymes that generate fattyacids and glycerol by fat digestion in aqueous media. Majority of industrial lipases are microbial ones, bacterial or fungal origin (Jaeger and Reetz 1998). Cellulases (EC 3.2.1.4) are a miscellany of endocellulases, exocellulases and cellobiose that generate cellulolysis the degradation of cellulose the most abundant carbon source in earth. Majority of cellulose in earth is considered as waste, but they can be the best source of food and potential source of energy (Elder et al., 1986 ). Xylanases (EC 3.2.1.8) are major hemicellulases that breaks down xylan a major plant cell wall material into xylose. Xylanases and cellulases are mainly of microbial origin and have also been reported from certain marine algae, snails, crustaceans, insects and seeds of plants (Walia et al., 2013 Chandra et al 2020).
Microbial protease, lipase, cellulase and xylanase are used in textile, pulp and paper, food and beverage and animal feed industries. They are also used in the production of biofuels, and eco-friendly bioconversion and management of waste materials and in pollution control (Liu and Kokare 2017, Subramaniyan and Prema 2002, Ramnath et al., 2017, Ali et al., 2016, Romdhane et al., 2010). Proteases and lipases have importance in medicinal and pharmaceutical sector (Andualema and Gessesse 2012, Singh et al., 2016). They are also used for degumming in silk and leather industry and for synthesis of peptides and esters. Proteases are made use in recovery of silver from photographic films and X-ray et al., (Ali et al., 2016). The ever increasing demand for microbial enzymes is because of the specific substrate specificity, low and high temperature and pH stability and optima (Singh et al., 2016, Treichel et al., 2010). For an enzyme to be industrially useful it must be thermo stable and must maintain activity even in the presence of solvents (Hasan et al., 2006). Enzymes which are fundamentally stable and active have more use in industrial sector. The following paper deals with the screening, production and characterisation of four industrially important enzymes, protease, lipase, cellulose and xylanase by Serratia marcescens strain VT 1.
MATERIAL AND METHODS
Microorganisms: The microorganism used in the present work was Serratia marcescens strain VT 1 (SMVT1) was isolated from oil contaminated soil collected from Althara Devi temple in Trivandrum district of Kerala. The organism was maintained by sub culturing in olive oil enriched nutrient agar plates (peptone – 0.5 g, yeast extract – 0.5 g, NaCl – 0.5 g, agar – 2 g & olive oil 1 ml for 100 ml). The cultures were incubated at 37°C for two days and were stored at 4°C in refrigerator.
Screening for enzymes – plate assay:Protease production:Casein agar plates composed of casein – 2 g, agar- 2 g, glucose – 0.5 g, NaCl – 0.5 g, peptone – 0.5 g & yeast extract – 0.5 g in 100 ml distilled water were streaked with microbial culture and kept in incubator for 2 days at 37°C. The plates were observed for clear zones around colonies which confirm protease activity.
Lipase production:Rhodamine olive oil agar plates were used to screen the presence of extracellular lipase (Castro-Ochoa et al., 2005). The media composed of peptone – 0.5 g, yeast extract – 0.5 g, NaCl – 0.5 g, agar – 2 g, olive oil – 3 ml & rhodamine – 0.1 mg for 100ml.The inoculated plates were incubated at 37°C for 48 h and observed under UV light for fluorescent orange halos around colonies which confirms lipase production.
Cellulase production:Screening test for cellulase was done by streaking the microbial culture on to CMC (Carboxy methyl cellulose) agar plates (CMC – 0.5 g, peptone – 0.5 g, yeast extract – 0.5 g & agar – 2 g for 100 ml). The plates were incubated for 48 h at 37°C. The plates were flooded with 1 M, 0.1% Congo red for 15 minutes and washed with 1M NaCl solution. Presence of clear zone confirms cellulase activity (Gohel et al., 2014).
Xylanase production:The bacterial culture was grown on xylan containing medium (peptone – 0.06g, yeast extract – 0.06 g, MgSO4 – 0.02 g, K2HPO4 – 0.1 g, xylan – 0.5 g & agar – 2 g for 100 ml) for 48 h at 37°C. Congo red (1M, 0.1 %) was used for staining and 1 M NaCl was used for destaining (Subramaniyan, 2012).Clear zone around the colony confirms presence of extracellular xylanase.
Fermentation studies:Medium for protease production:The pre- inoculums was raised at room temperature for 24 h at 120 rpm in 50 ml liquid medium composed of casein – 0.5 g, glucose – 0.5 g, NaCl – 0.5 g, peptone – 0.5 g & yeast extract – 0.5 g, CaCl2.H2O – 0.05 g, KH2PO4 – 0.02 g & MgSO4.7H2O – 0.05 g. 5 ml of this culture was transferred to production media of same composition and incubated at room temperature at 120 rpm for 96 h. pH of the medium was adjusted to 7 using Na2CO3 and the total volume of the medium was made up to 100 ml. Samples were collected at intervals of 24 h and centrifuged at 10,000 rpm for 15 minutes at 4°C, the supernatant was stored at 4°C in refrigerator and used as crude enzyme.
Medium for lipase production:The pre- inoculums was grown in 50 ml nutrient broth (yeast extract- 0.5 %, peptone- 0.5 % & NaCl- 0.5%) by transferring a loop full of day old microbial culture, incubated at room temperature at 120 rpm for 24 h. The production medium contains 1% olive oil in addition to pre- inoculum media, pH- 7 (Selvamohan et al., 2012). The culture conditions were exactly same. Samples were collected at a regular interval of 24 h for 4 days. Crude enzyme was obtained by centrifuging the culture to separate the cells at 10,000 rpm for 15 minutes at 4°C. The crude enzyme was stored at 4°C in refrigerator.
Protein estimation and pH determination:Cell protein was estimated according to standard procedure (Lowry et al., 1951). Bovine Serum Albumin (BSA) was used as standard. The pH values of the culture medium at different intervals were also noted.
Quantification of enzyme activity Lipase assay: Lipolytic activity was determined by using paranitrophenyl palmitate (PNPP) as substrate (Yagiz et al., 2007). Reaction mixture (9 ml of 50 Mm Tris HCl pH – 8 containing 40 mg triton X- 100 &10 mg gum arabic mixed with 3 mg of PNPP in 1 ml propane- 2- ol) with 0.1 ml of crude enzyme was incubated at 37°C for 30 minutes, and the release of p- nitrophenol was measured calorimetrically at 410 nm. The amount of enzyme required to release 1μmol of p- nitrophenol per minute per ml from PNPP was defined as the unit enzyme activity. P- nitrophenol was used as the standard.
Protease assay:Protease activity was measured using casein as substrate (Tsuchida et al., 1986). 0.5 ml of crude enzyme was mixed with0.5 ml of substrate (2% casein in sodium phosphate buffer pH- 7) and incubated at 40°C for 10 minutes. The reaction was terminated by adding 1 ml of 10% TCA. 0.5 ml of phosphate buffer with substrate and 1 ml TCA was used as blank. The resulting mixture was centrifuged at 2000 rpm for 5 minutes. To 1 ml of supernatant 5 ml of 0.44 M Na2CO3was added and incubated for 10 minutes. To this 2 fold diluted 0.5 ml Folin- Ciocalteau reagent was added and allowed to stand for 20 minutes; absorbance was measured at 660 nm. One unit of enzyme activity is defined as the amount of enzyme that released 1μmol of tyrosine per minute per ml. Bovine Serum Albumin (BSA) was used as standard.
Biochemical characterization :The biochemical characterization of crude enzyme was carried out with respect to optimum pH, pH stability, optimum temperature, temperature stability and effect of solvents.
Optimum pH and pH stability:Optimum pH for enzyme activity was determined at different pH values ranging from 4-11 by preparing substrates in appropriate buffers (protease: 4 & 5- sodium acetate buffer, 6 to 8- sodium phosphate buffer and 9 to 11- sodium carbonate buffer, lipase: 4 & 5- sodium acetate buffer, 6- sodium phosphate buffer, 7 to 9- Tris HCl and 10 & 11- sodium carbonate buffer). Enzyme assays were done according to the above mentioned procedures. The effect of pH on enzyme stability was studied by incubating the enzyme in desired buffer with optimum pH for two hours and calculating the residual activity for every half an hour.
Optimum temperature and temperature stability:The optimum temperature was determined by performing respective enzyme assay at different temperature ranging from 30- 80°C. Thermal stability was estimated by pre- incubating the crude enzyme at optimum temperature for two hours. Residual activity was determined for every half an hour.
Effect of solvents on enzyme activity: The different solvents used in the study ethanol, methanol, butanol, isopropanol, chloroform, ethyl acetate, acetone and hexane were selected based on their log P values. Crude enzymes were mixed with solvents to make a final concentration of 20% (v/v); the mixture was mixed well and stored at 4°C. The residual activity was determined after two hours of incubation by colorimetric method. Enzymes mixed with distilled water 20% (v/v) was used as control.
RESULTS AND DISCUSSION
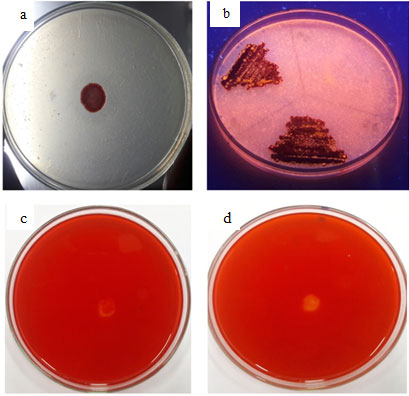
Figure 1: Plate Assay (A) Protease, (B) Lipase, (C) Cellulase And (D) Xylanase From SMVT1
S. marcescens VT 1 was subjected to screening by plate assays to confirm the presence of extracellular proteases, lipases, cellulases and xylanases (Fig: 1). After 48 h of incubation clear zone was visible around the bacterial colony in casein agar plates confirming protease activity. Hydrolysis of casein and gelatin was used as the preliminary test for isolation of protease producing Serratia sp RSPB11 (Bhargavi and Prakasham 2012). 14 protease producing strains were isolated on casein agar plates (Vakilwala and Patel 2017). Under UV light orange fluorescent halos were clearly visible resulting from the reaction of fatty acids and rhodamine pointing towards expression of extracellular lipase. Extracellular lipase production of Serratia marcescens and S. aureus, on Rhodamine B plates have been previously reported (Kouker and Jaeger 1987). Ejike and coworkers (2017) used Rhodamine B plates for preliminary screening of lipase producing organism S. marcescens. Majority of the lipases produced by microorganisms are extracellular inducible enzymes transmitted to outer surface (Ota et al., 1982). Clear zones were completely absent in CMC agar plates and xylan agar plates, so it can be inferred that cellulase and xylanase production is absent, therefore as the next part of work the production pattern and biochemical characterization of protease and lipase were studied.
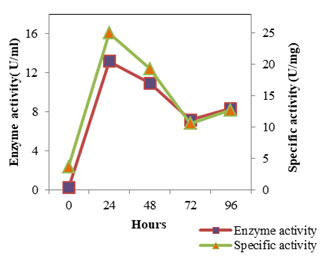
Figure 2: Enzyme Activity- SMVT1 protease
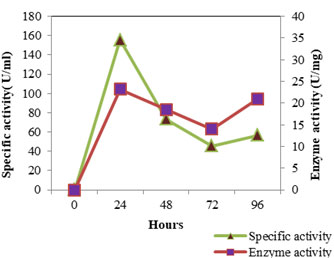
Figure 3: Enzyme Activity- SMVT1 LIPASE
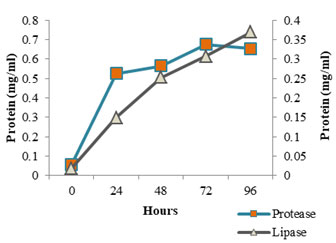
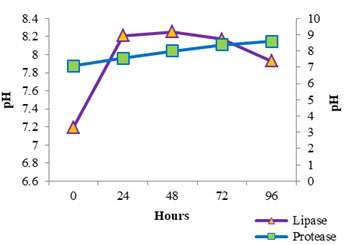
The enzyme production pattern was evaluated along with the change in pH profile of the medium and concentration of cell protein for a period of 96 h (Fig: 4 and 5). The extracellular protease production started with the log phase of growth and activity reached the peak value after 24 h (13.173 U/ml), thereafter the activity declined with time and showed a slight increase at 96 h (Fig: 2). The lipolytic activity of S. marcescens VT 1 was measured by colorimetry using PNPP as substrate at 420 nm. Study showed that the lipase activity was expressed maximum after 24 h of incubation, further a turn down in activity is seen which is followed by a rise in activity at 96 h (Fig: 3). The maximum activity recorded was 23.17 U/ml followed by 20.16 U/ml.
S. marcescens protease has been studied and described in previous papers (Henriette et al., 1993, Romero et. Al., 2001, Ustariz et. al., 2008). Excretion of extracellular protease of S. marcescens ATCC 25419 was reported to occur during the logarithmic growth phase and was determined to be highest during the stationary growth phase et al., et al., (Schmitz and Braun 1980). Lipid hydrolysis by S. marcescens was estimated and recorded earlier (Heller 1979, Prasad 2013, Abdou 2003). Henriette et al., (1993) based on their study reported that the S. marcescens released lipase during the stationary phase of growth after growing exponentially at 22°C for 6 h. Lipase production was found to begin after 5- 10 hours of growth at 27°C (Makhzoum et al., 1995). Proteolytic activity along with lipolytic activity of S. marcescens has been previously studied and recorded (Henriette et al., 1993, Begam et al., 2012, Abdou and Ohashi 1996). The rise in activity at 96 h may be due to the release of intracellular lipase into the medium due to death and lyses of bacterial cells. The pH of the protease medium was found to shift towards the alkaline side with time and growth of S. marcescens VT 1, while the pH of lipase medium turned alkaline first and later the alkalinity started to drop off after 48 h. The release of fatty acids, the byproduct of lipolysis into the medium could have resulted in reduction of alkalinity.
Figure 4: Cell Protein Profile Variation During Protease And Lipase Production By SMVT1.
Figure 5: Variation In Ph Values During SMVT1 Protease And Lipase Production.
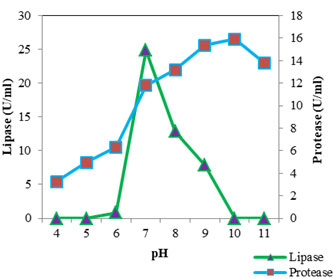
Figure 6: Optimum Ph Of Protease And Lipase
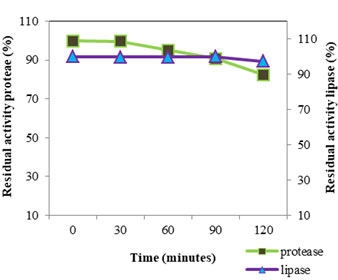
Figure 7: PH Stability Of Protease And Lipase At PH Value 10
The optimum pH (Fig: 6) for protease was found to be 10 (15.887 U/ml). At pH 9 the activity was found to be almost similar (15.346 U/ml). The proteolytic activity was found to decrease as the pH shifted towards acidic range. From this it is clear that the lipase of S. marcescens VT 1 is an alkaline protease and is active over a wide range of pH. Annapurna and colleagues (Annapurna et al., 2012) reported an alkaline protease form S. marcescens with optimum pH 10. S. marcescens metalloprotease SMP 6.1 with pH optimum of 10 was also reported (Salamone and Wodzinski 1997). Protease with an optimum pH of 8 and pH range of 6 to 11 was recorded in S. marcescens (Femi-Ola et al., 2014). Kim et al., (2007) reported the optimal pH of protease by S. marcescens as 7 using casein substrate. S. marcescens TKU019 protease exhibited a broad pH range 5-10 (Liang et al., 2010). The optimum activity of lipase was estimated at neutral pH. A sharp decline in activity was observed when pH shifted to 6 and activity was found to be absent at pH 5 and 4. Activity was found to decrease as the pH was raised and was lost at pH 10 and 11. S. marcescens lipase optimum pH was found to be 6.5 (Gao et al., 2004), 8 and retained 95% of maximum activity at 7 (Zaki and Saeed 2012) and 8-9 (Abdou 2003).
The pH stability (Fig: 7) of VT 1 protease was studied by incubating the crude enzyme in pH 10 for two hour at 4°C; assay was conducted every half an hour. The enzyme was found to be stable for 30 minutes in pH 10, after 60 minutes the enzyme retained 96.15% activity. After 2 h the enzyme lost 17.63% of its original activity. S. marcescens protease was found to be stable over a pH range of 5 to 10 under low temperature incubation and underwent inactivation in alkaline pH on incubation under elevated temperature (Miyata et al., 1970). Purified S. marcescens protease remained stable for 1 h at pH ranging from 6-9 and lost 40% of stability at pH 10 (Iqbal et al., 2018). For lipase pH stability study was conducted by incubating enzyme in Tris HCl buffer pH 7 and estimating activity every 30 minutes for two hours. VT 1 extracellular lipase was found to be stable for one and half hours and lost 2.73% of the original activity in the next 30 minutes and retained 97.27% activity by the end of two hours. S. marcescens lipase showed good stability between pH 6-9 (Makhzoum et al., 1995). Lipase from S. grimisii remained stable over the pH range 7-9 et al., (Abdou 2003). Abdou et al., (2003) found that purified S. marcescens lipase showed maximum stability at pH 8. In a study by Zaki and Saeed (2012) purified lipase from S. marcescens showed maximum stability at pH 8.
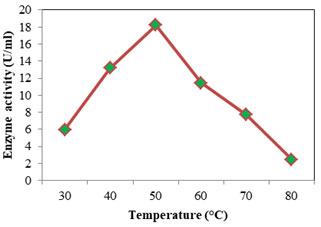
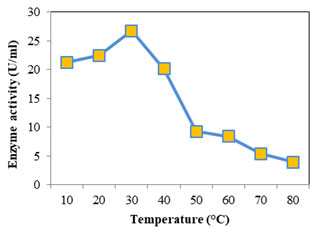
Optimum temperature for protease was assayed at temperature from 30 to 80°C; casein was used as the substrate at pH 7. Optimum activity recorded was 18.232 U/ml seen at 50°C (Fig: 8). At higher and lower temperatures the activity was found to decrease. For alkaline proteases the optimum temperature falls in the range 50- 70°C (Kumar and Takagi 1999). Femi-Ola and fellow workers (2014) obtained optimum proteolytic activity for S. marcescens at 50°C, at temperatures above and below reduction in activity was observed. Optimum activity was reported at 67°C for S. marcescens, alkaline protease (Annapurna et al., 2012). The optimum temperature of S. marcescens protease was found to be 50°C (Liang et al., 2010) and 42°C (Salamone and Wodzinski 1997). Optimum temperature for lipase (Fig: 9) was initially evaluated at temperature range 30- 80°C. Since the optimum activity was at 30°C (26.736 U/ml), lower temperatures 10 & 20°C were also considered. Optimum temperature for lipase from S. marcescens was determined as 35°C (Zaki and Saeed 2012).
S. marcescens lipase optimum temperature was reported to be 37°C (Abdou 2003), between 25- 35°C (Immanuel et al., 2008). With increase in temperature the lipolytic activity was found to decrease showing a sharp decline from 40- 50°C. At lower temperatures (30 to 10°C) the enzyme activity showed a slight decrease only, the activity was found to decrease by only 5.4 U/ml retaining almost 80% of the maximum activity. This shows the psychrophilic property of lipase. Abdou (2003) observed high lipase activity for S. marcescens at 5°C and low temperature activity of S. grimisii. Cold active enzymes have much economic and ecological benefit when compared with their equivalents which need high temperature to function (Marchi et al., 2007) Psychrophilic lipases have high catalytic activity and consume less energy at low temperatures which makes them efficient tools in the production of detergent, leather, food, pharmaceuticals, fine chemical and bioremediation (Joseph et al., 2008).
Figure 8: Optimum Temperature Of SMVT1 Protease
Figure 9: Optimum Temperature Of SM VT1 Lipase
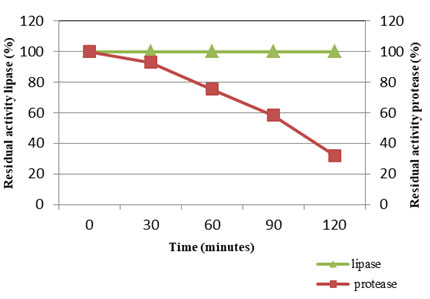
Temperature stability of VT 1 protease and lipase (Fig: 11) were studied at 50°C and 30°C (optimum temperature) respectively for a period of 2h and residual activity was determined after every half an hour. The VT 1 protease was active throughout the study period of two hours, but the activity was found to decrease drastically after 30 minutes. After 30 minutes of incubation at 50°C the activity decreased by 7%, thereafter the activity was found to lower by 61%. The lipase of S. marcescens was stable up to 45°C and lost 75% of stability at 60°C after 1 h of incubation (Iqbalet al., 2018). Matsumoto and partners (Matsumoto et al., 1984) showed the complete inactivation of S. marcescens protease above 65°C. Miyata et al., (1970) displayed the protease fro Serratia spp. to be relatively stable at 40°C throughout the 60 min of study, but mere heating for 15 min at 50°C resulted in complete inactivation. The lipase of VT 1 was thermo stable throughout two hours, showing that the lipase is highly stable at lower temperature. The S. marcescens lipase showed least residual activity at 85°C and was completely inactivated at 90°C. S. marcescens lipase was exhibited to be less thermostable compared to other psychrotroph lipases, at the same time is resistant to inactivation at lower temperatures (Abdou 2003). Thermal stability of S. marcescens lipase in the culture was showed to decrease with increasing temperatures (Gao et.al., 2004. After 24 h the residual activity was found to decrease by 61% and 36% after incubation at 35°C and 25°C.
Figure 10 Temperature Stability of SMVT1 Protease and Lipase.
Table 1. Solvent stability of SMVT1 protease and lipase
| Solvents | log P | Residual activity (%) protease | Residual activity (%) lipase |
| Methanol | -0.69 | 79.71 | 87.4 |
| Acetone | -0.61 | 54.73 | 85.23 |
| Ethanol | -0.18 | 82.28 | 76.55 |
| Isopropanol | 0.16 | 20.06 | 28.09 |
| Ethyl acetate | 0.71 | 38.83 | 37.63 |
| Butanol | 0.839 | 59.34 | 34.73 |
| Chloroform | 1.67 | 33.83 | 71.39 |
| Hexane | 3.769 | 65.79 | 92.67 |
Solvent stability of the enzymes were studied in eight different solvents with different log P values, which represents hydrophobicity level (Table: 1). Lower the log P value lower the hydrophobicity. Protease was found to be stable in polar solvents like methanol and ethanol, unlike the previously reported solvent tolerant protease from S. marcescens MH6 which displayed notable stability in hydrophobic solvents (Wan et al., 2010). Protease showed some stability in nonpolar solvent hexane also. Least amount of residual activity was observed in isopropanol and chloroform. Serratia sp. SYBC H protease retained over 90% activity even after 60 minutes of incubation at 40°C in 50% (v/v) hydrophilic organic solvents such as dimethylformamide, dimethylsulfoxide, and acetone (Li et al., 2010). The S. marcescens PPB-26 protease showed maximum stability in methanol and ethanol and was stable in all organic solvents except isopropanol (Thakur et al., 2016). The VT 1 lipase showed higher stability in methanol, acetone, and ethanol which are polar solvents with lower log P values, however maximum stability was observed in a nonpolar solvent hexane retaining 92% of activity after 2 h of incubation.
Lipases displaying stability in a wide group of organic solvents, regardless of their log P values have industrial importance in esterification, interesterification, and transesterification alias synthesis (Chakravorty et al., 2012). Serratia marcescens ECU1010 lipase presented commendable stability in many water miscible and immiscible solvents (Zhaoet al., 2008). Lipase from Pseudomonas reinekei displayed significant stability in hydrophilic solvents like ethanol and methanol (20% v/v) after 24 h of incubation (Priyanka et al., 2019). The activation of lipase in hydrophilic and hydrophobic solvents can be explained due to the amino acid solvent interactions leading to opening of the lid or flap covering the catalytic site keeping the enzyme in an open conformation (Bose and Keharia 2013, Cao et al., 2012 ) .
CONCLUSION
marcescens strain VT 1 a soil bacterium was isolated for the production of industrially important enzymes and was found to produce lipase and protease. A further characterization study showed the protease to have an optimum pH of 10 and was active over a wide pH range of 4-11 and showed stability at pH 10. Lipase had an optimum pH of 7 and was active in a pH range of 7-9 with good stability at pH 7 for 90 minutes. The optimum temperature of protease and lipase was found to be 50 and 30°C. Lipase was found to be cold active and incredibly stable at low temperatures. SMVT 1 protease was found to be stable in hydrophilic solvents like ethanol and methanol, while lipase showed stability in solvents with both low and high log P values. S. marcescens strain VT 1 protease and lipase can be of great importance in industries like food processing, biodiesel production, and waste management.
ACKNOWLEDGEMENTS
We would like to acknowledge the Principal, University College, Palayam, Thiruvananthapuram and Director, Department of Collegiate Education for all the facilities and assistance given during the period of work. VK is thankful to University of Kerala for the boundless research assistance.
REFERENCES
Abdou AM, Ohashi T (1996) Behavior of lipolytic and proteolytic Gram-negative psychrotrophic bacteria isolated from raw milk, cream and yoghurt in Egypt. J. Dairy Food Sci 45: A97-A104.
Abdou MA (2003) Purification and partial characterization of psychrotrophic Serratia marcescens lipase. J. Dairy Sci. 86: 127-132.
Ali N, Ullah N, Qasim M, Rahman H, Khan SN, Sadig, A, Adnan M (2016). Molecular characterization and growth optimization of halo-tolerant protease producing Bacillus subtilis Strain BLK-1.5 isolated from salt mines of Karak, Pakistan. Extremophiles 20: 395-402.
Andualema B, Gessesse A (2012) Microbial Lipases and Their Industrial Applications: Review. Article in Biotechnology (Faisalabad) 11(3): 100-118.
Annapurna SA, Singh A, Garg S, Kumar A, Kumar H (2012) Production and characterization of thermo tolerant alkaline protease from Serratia marcescens. Asian Jr. of Microbiol. Biotech. Env. Sc 14(4): 591-596.
Barrett AJ, McDonald JK (1986) Nomenclature: protease, proteinase and peptidase. Biochem J 237 (3): 935.
Beg QK, Gupta R (2003) Purification and characterization of an oxidation stable, thiol- dependent serine alkaline protease from Bacillus mojavensis. Enzyme microb Tecnol 32: 294-304.
Begam SM, Pradeep SF, Pradeep VB (2012) Production, purification, characterization and applications of lipase from Serratia marcescens MBB05. Asian J of Pharm Clin Res 5: 0974-2441.
Bhargavi PL, Prakasham RS (2012) Proteolytic enzyme production by isolated Serratia sp RSPB11: role of environmental parameters. Current Trends in Biotechnology and Pharmacy 6 (1): 55-65.
Bose H, Keharia (2013) Production, characterization and applications of organic solvent tolerant lipase by Pseudomonas aeruginosa AAU2, Biocatal. Agric. Biotechnol 2 (3): 255–266.
Cao Y, Zhuang Y, Yao C, Wu B, He B (2012) Purification and characterization of an organic solvent stable lipase from Pseudomonas stutzeri LC2-8 and its application for efficient resolution of (R, S)-1-Phenylethanol, Biochem. Eng. J. 64: 55–60.
Castro-Ochoa L, Rodriguez-Gomez C, Valerio-Alfaro G, Oliart Ros R (2005) Screening, purification and characterization of thermoalkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enzyme Microb Tech 37: 648-654.
Chakravorty D, Parameswaran S, Dubey VK, Patra S (2012) Unraveling the Rationale Behind Organic Solvent Stability of Lipases. Appl Biochem Biotechnol 167:439–461.
Chandra C Enespa, Ranjan Singh & Pankaj Kumar Arora (2020) Microbial lipases and their industrial applications: a comprehensive review Microbial Cell Factories BMC Springer Volume 19, Article number: 169 (2020)
Chapman J, Ismail AE, Dinu CZ (2018) Industrial Applications of Enzymes: Recent Advances, Techniques, and Outlooks. Catalysts 8: 238.
Eddehech, A., Zarai, Z., Aloui, F., Smichi, N., Noiriel, A., Abousalhamb, A., Gargouria, Y., (2019). Production, purification and biochemical characterization of a thermoactive, alkaline lipase from a newly isolated Serratia sp.W3 Tunisian strain. Int. J. of Biol. Macromol. 123, 792-800.
Ejike EN, Ejike BU, Onyeanula EO (2017) Influence of Environmental Parameters on Lipase Production by Serratia Marcesens. IOSR Journal of Biotechnology and Biochemistry 3(5): 28-33.
Elder, Chahal DS, Ishaque M (1986) Integrated processes for production of edible protein and fuel ethanol from biomass. Eutropic 22: 130-131.
Femi-Ola OT, Akinsanmi OP, Bamidele OS (2014) Production and Characterization of Protease from Serratia marcescens. AU J.T 18(1): 1-10.
Gao L, Xu, Li XJ, Liu ZZ (2004) Optimization of Serratia marcescens lipase production for enantioselective hydrolysis of 3-phenylglycidic acid ester. J Ind Microbiol Biotechnol 31: 525-530.
Gohel HR, Contractor CN, Ghosh SK, Braganza VJ (2014) A comparative study of various staining techniques for determination of extra cellular cellulase activity on Carboxy Methyl Cellulose (CMC) agar plates. Int.J.Curr.Microbiol.App.Sci 3(5): 261-266.
Hasan F, Shah A, Hameed A (2006) Industrial applications of microbial lipases. Enzyme Microb Technol 39: 235-251.
Heller BK (1979) Lipolytic activity copurified with the outer membrane of Serratia marcescens. J Bacteriol 140: 1120-1122.
Henriette C, Zinebi S, Aumaitre M, Petitdemange E, Petitdemange H (1993) Protease and lipase production by strain of Serratia marcescens (532S). J Ind Microbiol 12: 129-135.
Immanuel G, Esakkiraj P, Jebadhas A, Iyapparaj P, Palavesam A (2008) Investigation of lipase production by milk isolate Serratia rubidaea. Food Technol Biotechnol 46: 60-65.
Iqbal A, Al Hakim, Md Hossain S, Md Rahman R, Islam K, Md Azim F, Ahmed J, Md Assaduzzaman, Md Hoq M, Azad AK (2018) Partial purification and characterization of serine protease produced through fermentation of organic municipal solid wastes by Serratia marcescens A3 and Pseudomonas putida A2. Journal of Genetic Engineering and Biotechnology 16(1): 29-37.
Jaeger KE, Reetz MT (1998) Microbial lipase form versatile tools in Biotechnology. Trends in Biotechnology 16(9): 396-403.
Joseph B, Ramteke PW, Thomas G (2008) Cold active microbial lipases: some hot issues and recent developments. Biotechnol Adv 26: 457–470.
Kim HS, Golyshin PN, Timmis KN (2007) Characterization and role of a metalloprotease induced by chitin in Serratia sp. KCK. J Indus. Microbiol. & Biotech 34: 715-721.
Kouker G, Jaeger EK (1987) Specific and sensitive plate assay for bacterial lipases. Appl Environ Microbiol 53: 211-213.
Kumar CG, Takagi H (1999) Microbial alkaline protease: from a Bio industrial viewpoint. Biotechnol Adv 17: 561-594.
Li GY, Cai YJ, Liao XR, Yin J (2010) A novel nonionic surfactant- and solvent-stable alkaline serine protease from Serratia sp. SYBC H with duckweed as nitrogen source: production, purification, characteristics and application. Journal of Industrial Microbiology & Biotechnology 38: 845–853.
Liang TW, Kuo YH, Wu PC, Wang CL, Dzung NA, Wang SL (2010) Purification and characterization of a chitosanase and a protease by conversion of shrimp shell wastes fermented by Serratia marcescens Subsp. Sakuensis TKU019. J Chin. Chem So 57: 857-863.
Liu X, Kokare C (2017) Chapter 11 – Microbial Enzymes of Use in Industry. Biotechnology of Microbial Enzymes: Production, Biocatalysis and Industrial Applications. Academic press. 267-298.
Lowry OH, Rosebrough NJ, Farr LA, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265-275.
Makhzoum DA, Knapp JS, Owusu RK (1995) Factors affecting growth and extracelluar lipase production by Pseudomonas fluorescens. Food Microbiology 12, 277-290.
Marchi P, Longhi V, Zangrossi S, Gaetani E, Briani F, Deho G (2007) Autogenous regulation of Escherichia coli polynucleotide phosphorylase during cold acclimation by transcription termination and antitermination. Mol Genet Genom 278: 75-84.
Matsumoto K, Maeda H, Takata K, Kamata R, Okamura R (1984) Purification and Characterization of Four Proteases from a Clinical Isolate of Serratia marcescens kums 3958. Journal of bacteriology 157(1): 225-232.
Miyata K, Maejima K, Tomoda K, Isono M (1970) Serratia Protease Part I. Purification and General Properties of the Enzyme. Agr. Bioi. Chern. 34(2): 310-318.
Ota Y, Gomi K, Kato S, Sugiura T, Minoda Y (1982) Purification and some properties of cellbound lipase from Saccharomycopsis lipolytica. Agricultural and Biological Chemistry 46: 2885-2893.
Prasad MP (2013) Production of extracellular lipase by Serratia marcescens isolated from industrial effluent. Int J Curr Res Acad Rev1: 26-32.
Priyanka P , Kinsella G, Henehan GT, Ryan BJ (2019) Isolation, purification and characterization of a novel solvent stable lipase from Pseudomonas reinekei. Protein Expr Purif 153:121-130.
Ramnath L, Sithole B, Govinden R (2017) Identification of lipolytic enzymes isolated from bacteria indigenous to Eucalyptus wood species for application in the pulping industry. Biotechnology Reports 15: 114-124.
Rao CS, Satish T, Ravichandra P, Prakasham R (2009) Chatacterisation of thermo and detergent stable serine protease from isolated Bacillus circulans and evaluation of ecofriendly applications. Process Biochem 44: 262- 268.
Romdhane IB, Fendri A, Gargouri Y, Gargouri A, Belghith H (2010) A novel thermoactive and alkaline lipase from Talaromyces thermophilus fungus for use in laundry detergents. Biochemical Engineering Journal 53: 112-120.
Romero F. J, Garcia L.A, Salas J. A. Diaz M. Quirós L. M. (2001) Protease production from whey at high concentrations by Serratia marcescens. Process Biochemistry 36 (6), 507-515.
Salamone PR, Wodzinski RJ (1997) Production, purification and characterization of a 50-kDa extracellular metalloprotease from Serratia marcescens. Applied Microbiology and Biotechnology volume 48: 317-324.
Schmitz G, Braun V (1980) Excretion of a protease by Serratia marcescens. Arch Microbiology 124: 55-61.
Selvamohan T, Ramadas V, Sathya A (2012) Optimization of lipase enzyme activity produced by Bacillus amyloliquefaciens isolated from rock lobster Panlirus homaru. Int. J M. Eng. Res., 2: 4231-4234.
Singh R, Mittal A, Kumar M, Mehta PK (2016) Microbial Proteases in Commercial Applications. Journal of Pharmaceutical, Chemical and Biological Sciences 4(3): 365-374.
Subramaniyan S, Prema P (2002) Biotechnology of Microbial Xylanases: Enzymology, Molecular Biology, and Application. Critical Reviews in Biotechnology 22: 33-64.
Subramaniyan S. (2012) Isolation, Purification and Characterisation of Low Molecular Weight Xylanase from Bacillus pumilus SSP-34 Appl. Biochem & Biotech. 166(7):1831-42.
Thakur S, Sharma NK, Thakur N, Savitri, Bhalla TC (2016) Organic solvent tolerant metallo protease of novel isolate Serratia marcescens PPB-26: production and characterization. 3 Biotech 6(2):180.
Treichel H, Oliveira D, Mazutti MA, Luccio ML, Oliveira JV (2010) A Review on Microbial Lipases Production. Food Bioprocess Technol 3:182-196.
Tsuchida O, Yamagota Y, Ishizuka J (1986) An alkaline proteinase of an alkalophilic Bacillus sp. Curr Microbiol 14: 7-12.
Ustariz JF, Laca A, Garcia LA, Diaz M (2008) Fermentation conditions increasing protease production by Serratia marcescens in fresh whey . Rev. Tec. Ing. Univ. Zulia 31(1) 79- 89.
Vakilwala M, Patel D (2017) Isolation and screening of protease producing organisms from soil sample. Int. J. Res and Sci. Innovation 4(4): 75.
Walia A, Mehta P, Chauhan A and Shirkot CK (2013) Optimization of cellulase free xylanase production by alkalophilic Cellulosi microbium sp. CKMX1 in solid state fermentation of apple pomace using central composite design and response surface methodology. Ann Microbiol 63: 187- 198.
Wan, Hua M, Wu B, Ren W, He BF (2010) Screening, characterization, and cloning of a solvent-tolerant protease from Serratia marcescens MH6. J. Microbiol. Biotechnol 20(5): 881-888.
Yagiz F, Kazan D, Akin AN (2007) Biodiesel production from waste oils by using lipase immobilized on hydrotalcite and zeolites. Chem Eng J 134: 262-267.
Zaki HN, Saeed ES (2012) Production, Purification and characterization of extra cellular lipase from Serratia marcescens and its potential activity for hydrolysis of edible oils. J Al-Nahrain Univ 15: 94-102.
Zhao LL, Xu JH, Zhao J, Pan J, Wang ZL (2008) Biochemical properties and potential applications of an organic solvent-tolerant lipase isolated from Serratia marcescens ECU1010. Process Biochemistry 43(6): 626-633.