Xavier Research Foundation, Loyola Centre for Research & Development,
St Xavier’s College Campus, Navrangpura, Ahmedabad, Gujarat, India.
Corresponding Author Email Sudha.sahay@xrf.res.in,
Article Publishing History
Received: 11/07/2022
Accepted After Revision: 29/08/2022
The potential microalgal strains that are robust and display high growth and lipid accumulation rates are an important prerequisite for using them as a bioenergy source. We have isolated and screened six morphologically different microalgae strains, isolated from the Sabarmati River in Gujarat, India. The growth rates and lipid productivity of all six microalgae were assessed. Three potential microalgae strains were screened based on maximum biomass and lipid production. We named them MA001, MA002 and MA003. We identified MA001 as Micractinium reisseri based on its 28S rRNA sequencing. The M. reisseri showed an optimal growth rate of 2 g/L (dry weight) and 52 % lipid content after 20 days of cultivation in a normal artificial saline medium. Further analysis of lipid accumulation in M. reisseri was investigated at different concentrations of nitrogen. M. reisseri accumulated the highest amount of lipid under nitrogen starvation.
Artificial Saline Medium, Biodiesel, Bioenergy, Micractinium Reisseri, Microalgae.
Sahay S, Braganza V. Effect of Nitrogen on the Biomass Production and Lipid Accumulation of Micractinium Reisseri: A Potential Microalgal Strain for Bio-Fuel Production. Biosc.Biotech.Res.Comm. 2022;15(3).
Sahay S, Braganza V. Effect of Nitrogen on the Biomass Production and Lipid Accumulation of Micractinium Reisseri: A Potential Microalgal Strain for Bio-Fuel Production. Biosc.Biotech.Res.Comm. 2022;15(3). Available from: <a href=”https://bit.ly/3qjj52Z“>https://bit.ly/3qjj52Z</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Biofuel has caught substantial attention worldwide now-a-days as an alternative fuel due to its capability to adapt with gasoline for a maximum 85% blend without any engine modification. Subsequently, the suitability of microalgae for biofuel is being continuously queried by researchers and environmentalists (Hossain et al. 2017; Iqbal and Khan 2018; Hossain et al. 2019; Taofeeq et al. 2021). A well-managed and equipped microalgal bio-refinery can produce biodiesel and other value-added products such as protein, carbohydrates and a range of fatty acids (FAs) e.g., omega-3 fatty acids etc., and some non-nutritional value-added by-products such as bioplastic etc. (Beckstrom et al. 2020; Rajesh et al. 2020). However, the commercial production of microalgae biofuel is unfortunately offset (Chozhavendhan et al. 2022).
The oil content of the microalgae, the nitrogen content of the wastes, and the extraction and transesterification efficiencies significantly affect the biodiesel production in the integrated process, implies that improving these parameters is significant in increasing the feasibility of the integrated process (Deribew and Abubeker 2020; Zewdie and Ali 2020; Chozhavendhan et al. 2022). It was estimated that the current cost of producing 1 ton of microalgal biomass with an average 55% (w/w dry weight) oil content needs to be reduced by 10-fold in order to be competitive with petroleum diesel (Nazifa et al. 2021; Chandima and Judy 2022).
Therefore, photobioreactors and other effective methods have been designed for cultivation to enhance the rate of production of biomass (Patel et al. 2019; Zhang et al. 2022). However, one more factor, i.e., the potential of microalgae, cannot be ignored while improvising the production strategies. The high-yielding microalgae strain can directly translate into an overall increase in biodiesel production. Also, the lipid production during normal growth needs to be distinguished from lipid accumulation in response to adverse conditions e.g., nutrient starvation (Asma et al. 2022). The quantitative and qualitative differences in the lipid content of a given species affect the quality of biodiesel and its ability to meet fuel standards (Knothe 2005; Hu et al. 2008; Asma et al. 2022).
To date, research efforts have been focused on microalgal lipid production under several different growth conditions (Wagenen et al. 2012; Shafik et al. 2015, Reeza and Mallick 2021; Zhang et al. 2022). The direct comparison of various microalgae in different studies is unreliable because various researchers have used different growth conditions and experimental parameters for each species and also different methods have been used for lipid extraction (Zhang et al. 2022). However, the one that has a large accumulation of lipids proves to be a promising species for biodiesel production at a commercial level. Therefore, it is very important that one should have a potential microalgae strain along with a potential system of biodiesel production (Zhang et al. 2022).
In this study, few microalgae strains were isolated from Sabarmati River, Ahmedabad, Gujarat, India to screen the potential candidate for biomass and biofuel production. Total six microalgae strains were screened, of which three of them were shown to produce higher biomass and total lipid content. We named these three potential microalgae strains as MA001, MA002 & MA003. After various nutrient media screening, the artificial saline medium (ASM) was proved to be the most suitable medium for the cultivation of MA001, MA002 & MA003.
After 20 days of incubation MA001, MA002 & MA003 were assayed for the production of total biomass estimated as 0.2%, 0.1% and 0.04% (dry weight) and total lipid content was 52%, 47% and 65% (w/v) respectively. Further molecular identification by using 28S rRNA sequencing, identified the strain MA001 as Micractinium reisseri. Since, M. reisseri was found to produce higher biomass among all three screened potential strains; therefore, further studies were done to analyze the characteristics of M. reisseri. In this study the importance of nitrogen for the biomass production and lipid accumulation in M. reisseri are discussed.
MATERIAL AND METHODS
A total of six water samples (~2 L each) were collected from Vasna Barrage, Sabarmati Riverfront, Ahmedabad (Latitude 23.0080261 & Longitude 72.5716496) from 10 and 20 feet away from the river bank at three surfaces viz. surface water, 1 foot and 5 feet below the water surface. The temperature and pH of the collected water samples were noted on the spot at the time of collection. Isolation and purification of microalgal strains was done by serial dilution technique followed by plating of each dilution sample on four nutrient media viz., C10, B11, artificial saline medium (ASM) and artificial saline water (ASW) with 1% agar, pH 8.0 and were allowed to incubate under controlled environmental conditions in the growth room where the temperature was set at 25oC, light intensity of 3000 Lux, white LED light with 16 hours light and 8 hours dark photoperiod and 80% relative humidity (RH). A total of around 100 microalgal colonies along with the other microbial colonies were obtained. After several steps of isolation and purification, there were six green microalgal colonies with different morphology and green shades were selected.
Out of six selected colonies, one was successfully grown in B11 medium, the second was in C10 medium, the third was in ASW medium and the remaining three were grown in ASM medium. All six strains were shown to be axenic microalgae cultures. For further studies, these six axenic microalgae cultures were analyzed for their characteristics. Based on the estimated rate of growth and total oil content in all six selected microalgae strains, the three microalgae strains were screened for the most potential microalgal candidates and, therefore, selected for further studies. These three microalgae strains were labelled as MA001, MA002 & MA003 for which ASM medium was the most supportive nutrient medium.
A morphological study of the selected microalgae strains was done under the fluorescence microscope (Nikon) at 40x and 100x magnification. Growth parameters of the microalgae strains were measured every seven days of incubation up to 70 days. The cell density was measured using a hemocytometer. The microalgae biomass in a 10 ml liquid culture was measured by centrifugation at 4000 rpm for 30 min. The whole set of experiments was done in triplicate. The data obtained at different incubation periods was recorded.
For the molecular identification of microalgae strain 001, the fresh sample was harvested in 10 mL of liquid culture by centrifugation. Microalgae sample was washed with sterile Millipore water and dried at room temperature (~30°C) for a few hours to remove excess liquid. The dried microalgae MA001 sample was subsequently for DNA extraction. The air-dried microalgae cells (1 g) were ground to a powder in a pre-chilled mortar and pestle. The powder was then transferred into a 2 µl microcentrifuge tubes.
Then 500 µl of the extraction buffer (CTAB-3% (w/v); Tris HCl-100mM; NaCl-2M; EDTA-25mM; Mercaptoethanol-4% (v/v); PVP-5% (w/v); pH 8.0) was added into it and mixed well. The mixture was incubated for 30 minutes at 70°C with periodic shaking. After incubation, the mixture was cooled at room temperature and then 500 µl chloroform: isoamyl alcohol (24:1) was added and mixed gently for 15 min. The mixture was centrifuged at 12000 rpm for 15 min. The above aqueous phase (about 500 µl) was transferred to a new tube and then added ice-cold isopropanol (700 ml).
The whole content was mixed gently and kept at -20oC for 10 min. Then, centrifuged the tubes at 10,000 rpm for 5 min. The supernatant was discarded. Cold 80% ethanol was added to the pellet to wash it twice by centrifugation at 10,000 rpm for 5 min. The ethanol was discarded, and the pellet was air dried. The pellet was re-suspended in 50 µl 1X TE buffer (10mM Tris HCl; 1Mm EDTA; pH 8.0) and stored at -20oC. DNA concentration and purity were determined by measuring the optical density using a UV- spectrophotometer (SHIMATZU).
For this, 10 µl of DNA sample was diluted with 100 µl of 1X TE buffer (10 X dilution), and the optical density was measured at 260 nm in a UV-Spectrophotometer (SHIMADZU) against 1X TE buffer. The DNA concentration was then calculated according to the standard method. Optical density (OD) was also taken at 280 nm (corresponding to protein), 230 nm (corresponding to RNA) and 320 nm (contamination). Total DNA purity was tested by a ratio of OD values at 230:260:280. Species identification of the microalgae strain MA001 was done by the molecular sequencing of the 28S rRNA subunit.
The primer sequence used for the PCR reaction was:
F- “ACCCGCTGAATTTAAGCATA” and R- “CCTTGGTCCGTGTTTCAAGA”
The produced DNA sequence was analyzed using BLAST to confirm the sequence identification.
Four sets of ASM media supplemented with three different concentrations of nitrogen compound were used to cultivate the microalgae strain MA001. ASM medium without nitrogen supplement was used as a control. In ASM medium, three different concentrations of sodium nitrate (NaNO3) supplement were used: 1.25, 2.5, and 5.0 g/L. The microalgae strain, MA001 with a cell density of 6.58 x 105 cells/ml was inoculated in all four sets of ASM medium containing 0, 1.25, 2.5 and 5.0 g/L concentrations of NaNO3. The microalgae inoculum and ASM medium ratio were maintained at 1:100 taken in 250 ml Erlenmeyer flasks. All inoculated flasks were incubated in static condition in a controlled environment at a temperature 25oC, a light intensity of 3000 Lux, white LED light with a photoperiod 16 hours of light and 8 hours of darkness, and a relative humidity (RH) of 70-80 %.
Growth parameters and biochemical assays of the growing microalgae strain were done on the 5th, 10th, 25th and 40th days of incubation. A 10 ml liquid microalgal culture was harvested from each flask and used to study the growth parameters such as (i) cell density; (ii) wet biomass; (iii) dry biomass; (iv) chlorophyll content; (v) total lipids and (vi) neutral lipids by using standard methods. Cell density in 1 ml of algal suspension culture was measured using a hemocytometer and optical density (OD650) was taken using an Ultraviolet-Visible spectrophotometer (SHIMADZU). All measurement were done in triplicates by taking algal culture from three parallel flasks of the same test sample. The linear correlation between the cell density of M. reisseri and OD650 was established for the subsequent M. reisseri density determination using the equation given by Li et al. (2010). The software EXCEL was applied to process the data, draw the scatter plot and calculate the correlation coefficient.
The linear correlation between OD650 and cell density was determined by the equation: D (x106 cells/ml) = 79.311x+70957; (R2 = 0.997). Wet biomass was determined by the centrifugation method. First weighed the empty centrifuge tube (15 ml volume). 10 ml of microalgae culture was taken in each centrifuge tube and centrifuged at 4000 rpm for 30 min. After centrifugation, the supernatant was decanted and again weighed the centrifuge tube with the microalgae pellet. The weight difference of centrifuge tubes before and after centrifugation was recorded. The centrifuge tubes with wet microalgae pellets were allowed to air-dry for 24 hours. The centrifuge tubes with dry microalgae pellets were weighed again. The weight difference between empty centrifuge tubes and the same centrifuge tube with an air-dried microalgae pellet after centrifugation was recorded.
For chlorophyll estimation the microalgae cells were harvested from a 10 ml suspension culture and washed with sterile Millipore water. Microalgal cells were partially dried at room temperature and then 10 ml of methanol was added to them. The cells were mixed thoroughly with the help of a glass rod, and then the content was transferred to an Erlenmeyer flask. The flask was closed tightly with aluminum foil and kept in a water bath at 60oC for 30 min. Then, it was allowed to cool at room temperature.
Methanol content was decanted in a 15 ml centrifuge tube. The final volume was adjusted to 10 ml by adding methanol. The tube was centrifuged at 8000 rpm for 10 min at ambient temperature. The supernatant was separated and the optical density (OD) of the supernatant was measured at the wavelengths of 663 nm and 645 nm in the UV-Spectrophotometer (SHIMADZU). Methanol was used as a control. The obtained values were used in the formulas, described by Gu et al. (2016) for the estimation of photosynthetic pigments.
Chlorophyll a (mg/g) = (12.7 x OD663) − (2.59 x OD645);
Chlorophyll b (mg/g) = (22.9 x OD645) − (4.7 x OD663);
Chlorophyll total (mg/g) = (8.2 x OD663) + (20.2 x OD645).
The Nile red staining was used to determine the lipid globular structures inside the microalgae cells as described by researchers (Matsunaga et al. 2009; Elumalai et al. 2011). The cells of microalgae strain MA001 were isolated from 1 ml suspension cultures in ASM media on the 40th day of incubation. The culture was centrifuged at 1,500 rpm for 10 min and sediment was washed with saline solution (0.5 ml) 2-3 times. The collected microalgal cells were suspended in 0.5 ml of Nile red solution (0.1 mg of Nile red/ml of acetone) and incubated for 10 min at room temperature.
The stained microalgal cells were washed with sterile reverse osmosis (RO) water, and the intracellular lipid content was observed under a fluorescent microscope (NIKON ECLIPSE E200). The neutral lipid content of MA001 at three different concentrations of nitrogen and control samples were determined by modified fluorescence protocol (Chen et al. 2009). 1 ml of microalgae suspension culture was taken in a microcentrifuge tube. Similarly, 1ml of sterile reverse osmosis (RO) water was taken in another microcentrifuge tube that was used as a blank. 50 µl DMSO (dimethyl sulfoxide) was added to all the tubes and mixed well. The tubes were kept in a water bath at 40oC for 1 min.
Tubes were cooled at room temperature and 10 µl Nile red stock (125 µg of Nile red/ml in acetone) was added. After a thorough mixing, the tubes were again kept in the water bath at 40oC for 30 min. with an intermediate shake. After algal cells were stained, fluorescence emissions were recorded with the Elisa Reader technique equipped with a 96-well plate reading mode. Analysis for all the samples was performed in triplicates. Fluorescence emissions were recorded for the stained microalgal cells at excitation and emission wavelengths of 490 nm and 580 nm, respectively, by using an ELISA Reader (Thermo Scientific). Observations were made at 10, 25 and 40 days of incubation. Total lipid was measured by the method Bigogno, et al. (2002). 100 mg of dry microalgae biomass was taken in a glass vial and 5 ml solvent I (10% DMSO in methanol) was added into it. The vial caps were tightly closed and placed in a 45oC water bath with occasional shaking for 45 minutes.
The vials were then cooled down to room temperature. 5 ml of solvent II (diethyl ether: hexane (1:1)) was added to it and it was placed in a 45oC water bath for 60 minutes. After cooling at room temperature, the content was transferred into 15 ml centrifuge tubes. Tubes were centrifuged at 5000 rpm for 10 min. The supernatant was transferred into a separating funnel. 10 ml of sterile distilled water was added to it. The content was mixed thoroughly and allowed to settle down for one night. Two solvent phases were separated. The upper organic phase, consisting mainly of lipids was collected in a pre-weighed vial. Then weighed the organic lipid content obtained from 100 mg of dry microalgae biomass.
RESULTS AND DISCUSSION
Light to dark green colonies were formed after about 10-20 days of incubation (Figure 1a). Six randomly selected colonies were observed under the light microscope. All colonies were shown to have common characteristics of the family Chlorophyceae. The size of the individual cells of each strain was in the range of 8-10 μm. All six colonies were shown to have different shades of light to dark green, which can be seen with the naked eye. The shape of all six types of cells varies from oval to spherical to spindle; each cell with a single nucleus as shown in figure 1(b-g). Strains MA001, MA002 and MA003 were grown successfully in ASM medium for 70 days in 100 mL Erlenmeyer flasks (Figure 1h). They were further cultivated in big glass jars with a 5 liters capacity in the ASM medium (Figure 1i).
There was no significant difference in the growth patterns that were observed even with an increase in the volume of ASM medium from 250 ml to 2500 ml. The ratio of inoculum to nutrient media was fixed at 1:100. The morphological characteristics of MA001, MA002 and MA003 are shown in table 1. After 20 days of incubation of microalgae strains MA001, MA002 & MA003, the estimated recovery of biomass was 2 g/L, 1 g/L and 0.4 g/L (dry weight) and lipid content was 52%, 47% and 65% respectively. The results indicated that biomass production was highest in the MA001 strain and lipid production was highest in the MA003 strain among all three.
Molecular identification was done for the strain MA001. Genomic DNA of MA001 was sequenced for the 28s RNA subunit from outsourcing (Eurofins). The nucleotide sequence of a small DNA fragment (~400 bp) is shown in figure 2. NCBI-Blast search of the sequence of a small DNA fragment of MA001 showed 100% similarity with microalgae – Micractinium reisseri belong to the family Chlorophyceae.
Figure 1: (a) Microalgae colonies on media plates; (b-g) Light microscopic images of six isolated
microalgal strains; (h) axenic microalgae cultivation in flasks and (i) jars.
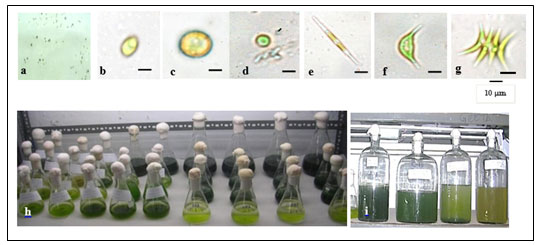
Table 1. Characteristics of 20 days old microalgae strains
| Parameter | MA001 | MA002 | MA003 |
| Cell Morphology | Dark green, oval and round, small, always dispersed. | Light green, round, bigger than MA001, sometimes in group or dispersed. | Light green, sickle shaped, pointed ends, flexible, always dispersed. |
| Dry Biomass ( g/L) | 2 | 1 | 0.4 |
| Lipid content (%) | 52 | 47 | 65 |
| De-oiled cake (g) | 1.2 | 1.1 | 0.45 |
Figure 2: 28S rRNA sequence of MA001 DNA fragment.

The effect of nitrogen on the growth and development of M. reisseri was studied under controlled environmental conditions. In general, the ASM medium contains 2.5 g/l NaNO3 as a nitrogen supplement, which we considered a normal concentration (1N2). The remaining two concentrations were half (1/2N2) and double (2N2) the normal nitrogen supplement concentration. ASM media without any nitrogen supplement was used as a control (-N2). M. reisseri was harvested on the 5, 10, 25 and 40 days of incubation. The growth parameters of M. reisseri were determined after each harvest. Cell density or the biomass productivity of M. reisseri was increased from low to high concentrations of nitrogen. Cell growth was also observed in control.
Figure 3: Cell Density of M. reisseri cultivated in ASM medium with N2 absent, 1.25 g/L (N2 half), 2,5 g/L (N2 full) and 5.0 g/L (N2 double) concentrations.
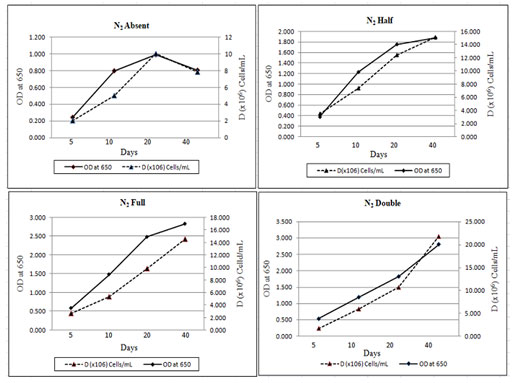
Figure 3 shows the total cell biomass with respect to the linear growth curve of M. reisseri with increasing concentrations of nitrogen. Observations were made for 40 days of incubation. It was observed that without nitrogen compound, there was no significant increase in the cell density. However, with an increase in nitrogen concentration in the ASM medium, there was a significant increase in the cell density with an increase in incubation period. A drastic increase in cell density was observed on the 40th day of incubation when there was the highest concentration of nitrogen (5 g/L).
The growth rate and cell density of M. reisseri grown on ASM medium with and without nitrogen supplement demonstrated that biologically available nitrogen support enhances CO2 fixation for photosynthesis, which brings about the cell growth. However, with a nitrogen supplement, the cell growth is further enhanced. The highest level of chlorophyll-a in 2N2 concentration was observed, followed by a decreasing order of chlorophyll-a content with decreasing concentrations of nitrogen, viz., 1N2, 1/2N2, 0N2 was observed up to the 40th day of incubation (Figure 4). However, the estimated amount of chlorophyll a in 1N2 was very close to that in 2N2. The medium without nitrogen supplement (-N2) contained the least chlorophyll-a of all (Reeza and Mallick 2021; Zhang et al. 2022).
Figure 4: Chlorophyll-a estimated in M. reisseri grown at four different concentrations of nitrogen in ASM media.
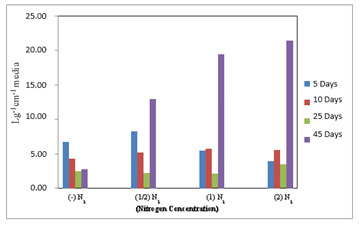
There was a marginal variation in the level of chlorophyll-b content in M. reisseri when grown at four different concentrations of nitrogen up to 25 days after inoculation (Figure 5). However, on the 40th day of incubation, there was a sudden increase in the level of chlorophyll b observed in all three concentrations of nitrogen. The highest amount of chlorophyll-b was observed in 2N2 followed by less in 1N2 and least in 1/2N2. M. reisseri growing in ASM medium without any nitrogen supplement (–N2) determined a negligible amount of chlorophyll-b. In all concentrations of nitrogen there was a slight decrease in the total chlorophyll up to 25 days, but it suddenly increased on the 40th day of incubation. (Figure 6).
Figure 5: Chlorophyll-b estimated in M. reisseri grown at four different concentrations of nitrogen in ASM media.
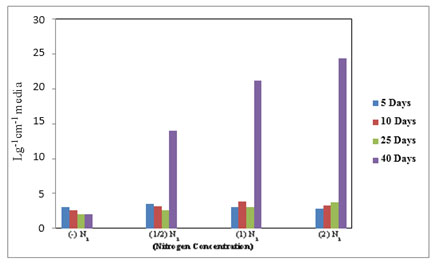
Figure 6. Total chlorophyll estimated in M reisseri grown at four different concentrations of nitrogen in ASM media.
It has been observed that when the nitrogen supplement was absent or of less amount in the ASM medium, there was the least production of total chlorophyll in the M. reisseri. Photochemical measurements show that nitrogen supplementation does not inhibit photosynthesis in phytoplankton. However, the rate of Photosystem-II re-oxidation becomes faster and photosynthetic electron transport is enhanced during growth on a high level of NaNO3 and cells maintain overall photosynthetic efficiency. As a result, the higher the level of chlorophyll, the greater the growth and development of microalgal cells.
The selection of solvent is very important in the pigment extraction process because the polarity of the solvent used for the extraction process plays a major role. In the present work, the pigment extraction with methanol was nearly completed in M. reisseri cells. Many factors, such as the cell wall structures of M. reisseri, a well performed homogenization and a complete extraction, facilitated the isolation of the pigments. It was suggested in earlier studies that the chlorophyll-a level was found to be the same in all microalgae groups (Chiu et al. 2009; Nazifa et al. 2021).
However, we have found that the level of chlorophyll a in M. reisseri varies at different concentrations of nitrogen. With an increase in nitrogen, the level of chlorophyll-a was increased due to the increase in cell growth. In other words, the amount of chlorophyll a pigment was directly proportional to the cell biomass at a specific nitrogen concentration. In a previous study, it was reported that the level of chlorophyll is found higher in the microalgae present in fresh water and in an environment where much light and temperature stratification are not seen (Li et al. 2010).
It was also determined that there were changes at the pigment level of algae species that live in an environment where light stratification is seen (Daphne et al. 2021). That is why one of the opinions is that the reason for the chlorophyll-a level to be higher with an increase in time of incubation and nitrogen concentration is linked with light and temperature stratification during the course of time when the samples were taken. Li et al. (2008) reported that the accumulation of CaCO3 was found to be very high on the cell walls of almost all microalgae, e.g., Codium tomentosum and Cladostephus verticillatus, which hinders the extraction process (Chandima and Judy 2022).
In our experiment, cell homogenization was a difficult step that might be due to CaCO3 accumulation, and therefore, all pigments were not extracted completely. As a result, chlorophyll-b was determined to be less in concentration as compared to chlorophyll-a in M. reisseri. An optimal protocol for ELISA was successfully applied to M. reisseri for lipid determination. We have used Nile red and DMSO (dimethyl sulfoxide) solutions to improve the effectiveness and efficiency of lipid fluorescence.
The microalgal extracts contained pigments that quenched Nile red fluorescence. Therefore, DMSO acts as a mild bleach solution to destroy pigments. DMSO was also introduced to microalgal samples as the stain carrier at an elevated temperature (Chen et al. 2009). After the algal cells were stained with Nile red and DMSO, the fluorescence was measured on a 96-well-microplate reader (ELISA) using medium scan control and high PMT detector voltage mode at the excitation and emission of 490 nm and 580 nm.
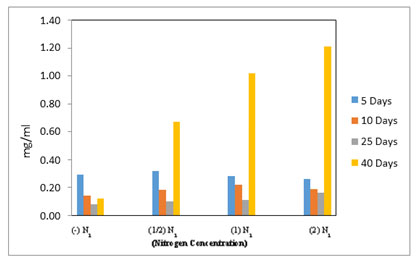
All M. reisseri cells contain measurable amounts of neutral lipids in the normal growth state. The observed fluorescence is not the result of inherent cellular fluorescence from chlorophyll as we have measured untreated M. reisseri suspension cultures without Nile red dye (control) as well as the ASM medium containing Nile red alone as an auto florescence. The amount of florescence is considered as an indication of the lipids. In table 1, we have mentioned that the total lipid content (polar and non-polar) in M. reisseri is 52 % of the total cell biomass (dry weight).
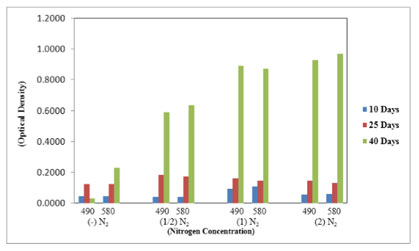
In the ELISA reader, the lipid globular molecules were reacted with Nile red and gave a yellow florescence. Figure 8 shows the increase in optical density of M. reisseri suspension at 490 and 580 nm with an increase in nitrogen concentration of ASM medium. This is because the increasing nitrogen concentration in the nutrient medium would decrease the lipid accumulation in the microalgal cells. Due to the least availability of lipid molecules, more of the Nile red dye would retain its red colour and consequently increase the optical density with respect to control.
Nile red (9-(Diethylamino)-5H benzo [α] phenoxazin-5-one) staining is specifically used to identify the intracellular lipid droplets from the biological samples (Greenspan et al. 1985). The results indicated that M. reisseri could not be affected by Nile red staining since oil droplets were not clear and the whole cells were stained red. Similar results were reported earlier in a few other green isolates. Sayeda et al. (2013) reported that certain blue green isolates were affected by the dye, including Microcystis aeruginosa and Phormidium rimosum where the yellow stained parts were clear. However, Chroococcus turgidus, Oscillatoria limnetica and Oscillatoria limosa cells showed yellow florescent colour under a florescent microscope even without adding the dye. So, this gives false results when referring to lipid content.
Figure 7. Total lipids in M. reisseri grown at four different concentrations of nitrogen in ASM
media (Optical density was measured at 490 and 580 nm).
The florescent method has been successfully applied to the determination of lipids in certain microalgae, but has been unsuccessful in many others, particularly those with thick, rigid cell walls that prevent the penetration of the dye (Held 2011). Judith et al. (2015) also mentioned that many of the microalgae strains do not give yellow colour with Nile red for lipid test. Indeed, interactions of the dye with proteins and/or other cellular components in cytosolic cells are major delay factors for colour development.
He explained that fluorescence and staining kinetics depend on the microalgae species and the size of lipid droplets as well as on the amount of the latter. Li et al. (2008) reported that increasing the stress in the growth conditions can increase lipid accumulation in microalgal cells. Nitrogen stress is one of them. There are reports that confirm the higher proportions of lipids (up to 60%) in microalgae obtained under nitrogen starvation (Bigogno 2000; Christos et al. 2020).
The fatty acid composition and fatty acid content changed with the ageing of the culture. Consequently, the total lipid content increased dramatically with an increase in the age of microalgae, at least up to 30 to 40 days. However, the non-polar or neutral lipid accumulation is an important aspect in terms of its biofuel production. Certain microalgal species can be induced to synthesize and accumulate extremely high proportions of triacyleglycerate (TAG). Stewart and Coyne (2011) describe chimeric enzymes that are involved in maintaining cell growth in the presence of nitrogen (Chandima and Judy 2022).
CONCLUSION
The findings of the present study has presented the potential to produce biodiesel from the microalgae M. reisseri – a model for neutral lipid production. The strength of M. reisseri is to predict with various morphological characteristics, developed at high nitrogen limitation to starvation. Both chlorophyll a and b were present in M. reisseri. The compositional information and methodology presented in this paper are valuable references for future studies on microalgae strain selection and algae cultivation, optimization, as well as for the production of biodiesel from microalgae.
ACKNOWLEDGEMENTS
The authors are thankful to the Xavier Research Foundation for supporting the studies under in-house R&D grant.
Data Availability Statement: All data / results / information is available with the authors and can be shared on a reasonable request made to the corresponding author when required.
Conflict of Interests: Authors declare no conflict of interests to disclose.
REFERENCES
Asma S, Seham M H, Ahmed I O, et al. (2022). Algal biomass valorization for biofuel production and carbon sequestration: a review. Environmental Chemistry Letters.
Beckstrom B D, Wilson M H, Crocker M, et al. (2020). Bioplastic feedstock production from microalgae with fuel co-products: A techno-economic and life cycle impact assessment. Algal Res Vol 46 No 101769.
Bigogno C (2000). Biosynthesis of Arachidonic Acid (AA) in the Microalga Parietochloris incise and the effect of environmental conditions on the production of AA. Ph.D. thesis, Ben Gurion University, Israel.
Bigogno C, Khozin-Goldberg I, Boussiba S, et al. (2002). Lipid and fatty acid composition of the green oleaginous alga Parietochloris incisa, the richest plant source of arachidonic acid. Phytochemistry Vol 60 No 5 Pages 497-503.
Chandima G and Judy M (2022). Development of Microalgae-based Biofuels as a Viable Green Energy Source: Challenges and Future Perspectives. Bio interface Research in Applied Chemistry Vol 12 No 3 Pages 3849-3882.
Chen W, Zhang C, Song L, et al. (2009). A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J Microbiol Methods Vol 77 No 1 Page 41.
Chiu S Y, Kao C Y, Tsai M T, et al. (2009). Lipid accumulation and CO2 utilization of Nannochloropsis oculata in response to CO2 aeration. Bioresour Technol. Vol 100 Page 833.
Chozhavendhan S, Karthigadevi G, Praveenkumar R, et al. (2022). Chapter 9 – Potentials and challenges in biodiesel production from algae—technological outlook. Biofuels and Bioenergy: Opportunities and Challenges, Pages 183-203.
Christos L, Jasper V H and Klaas R T (2020) .The Effect of Nitrogen Starvation on Biomass Yield and Biochemical Constituents of Rhodomonas sp. Front. Mar. Sci. Vol 7 No 563333.
Daphne D, Evanthia M, Daniel F, et al. (2021). Stratification strength and light climate explain variation in chlorophyll a at the continental scale in a European multilake survey in a heatwave summer. Limnology and Oceanography Vol 16 No 12 Pages 4314-4333.
Deribew T Z and Abubeker Y A (2020). Cultivation of microalgae for biofuel production: coupling with sugarcane processing factories. Energy, Sustainability and Society Vol 10 Page 27.
Elumalai S, Prakasam V, Selvarajan R, et al. (2011). Optimization of abiotic conditions suitable for the production of biodiesel from Chlorella vulgaris. Indian J. of Sci. & Tech. Vol 4 No 2 Page 91.
Greenspan P, Mayer E P, Fowler S D, (1985). Nile Red – a selective fluorescent stain for intracellular lipid droplets. The J of cell boil. Vol 100 Page 965.
Held P (2011). Monitoring algal cell growth using absorbance and fluorescence. In: BioTek Application.
Hossain N, Zaini J H and Mahlia T M (2017). A review of bioethanol production from plant-based waste biomass by yeast fermentation. Int J Technol. Vol 8 Pages 5–18.
Hossain N, Mahlia T M, Zaini J H, et al. (2019). Techno-economics and sensitivity analysis of microalgae as commercial feedstock for bioethanol production. In: Environmental progress and sustainable energy. New Jersey: John Wiley & Sons. Pages 1–14.
Hu Q, Sommerfeld M, Jarvis E, et al. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. Vol 54 Pages 621–639.
Iqbal M and Khan F U (2018). Hybrid vibration and wind energy harvesting using combined piezoelectric and electromagnetic conversion for bridge health monitoring applications. Energy Convers Manag. Vol 172 Pages 611–618.
Judith R, Hubert B, Bruno S J, et al. (2015). The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae. Biotechnology for Biofuels Vol 8 Page 42.
Knothe G (2005). Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Processing Technology Vol 86 Page 1059.
Li X, Hu H Y and Gan K (2010). Effect of different nitrogen and phosphorus concentration on growth, nutrient uptake and lipid accumulation of a freshwater microalgae Scenedesmus sp. Bioresour. Technol. Vol 101 Page 5494.
Li Y Q, Horsman M, Wang B, et al. (2008). Effects of nitrogen sources on cell growth and lipid accumulation of green algae Neochloris oleoabundans. Appl. Microbio. Biotech. Vol 81 Page 629.
Matsunaga T, Matsumoto M and Maeda Y (2009). Characterization of marine microalgae, Scenedesmus sp. Strain JPCCGA0024 toward biofuel production. Biotech. Let. Vol 31 Page 1367.
Nazifa R, Shams F A, Irfan A B, et al. (2021). Strategies to produce cost effective third-generation biofuel from microalgae. Front. Energy Res. Vol 9 Article 749968.
Patel A K, Joun J M, Hong M E, et al. (2019). Effect of light conditions on mixotrophic cultivation of green microalgae. Bioresour. Technol. Vol 282 Pages 245–253.
Rajesh B, Preethia J, Kavitha S, et al. (2020). Microalgae based biorefinery promoting circular bioeconomy-techno economic and life-cycle analysis. Bioresour. Technol. Vol 302 Pages 122822.
Reeza P and Nirupama M (2021). Microalgal biodiesel production: realizing the sustainability index. Frontiers in Bioengineering and Biotechnology Vol 9 Article 620777.
Sayeda M A, Entesar A, Sanaa A E, et al. (2013). Growth rate and fatty acids profile of 19 microalgal strains isolated from river Nile for biodiesel production. J. Algal Biomass Utln. Vol 4 No 4 Pages 51–59.
Shafik H M, Saad M G and El-Serehy H A (2015). Impact of nitrogen regime on fatty acid profiles of Desmodesmus quadricaudatus and Chlorella sp. and ability to produce biofuel. Acta Bot. Hung. Vol 57 Pages 205–218.
Stewart J J and Coyne K J (2011). Analysis of raphidophyte assimilatory nitrate reductase reveals unique domain architecture incorporating a 2/2 hemoglobin. Plant Mol. Biol. Vol 77 Page 565.
Taofeeq D M, Gusman N and Mahmud F (2021). Microalgae biofuels production: A systematic review on socioeconomic prospects of microalgae biofuels and policy implications. Environmental Challenges, Vol. 5 Article 100207 Pages 1-13.
Wagenen V J, Miller T W, Hobbs S, et al. (2012). Effects of light and temperature on fatty acid production in Nannochloropsis salina. Energies Vol 5 Pages 731–740.
Zewdie D T and Ali A Y (2020). Cultivation of microalgae for biofuel production: coupling with sugarcane processing factories. Energy, Sustainability and Society Vol 10 Page 27.
Zhang S, Zhang L, Xu G, et al. (2022). A review on biodiesel production from microalgae: Influencing parameters and recent advanced technologies. Front. Microbiol. Vol 13 Article 970028.


