1Department of Zoology, Jagadamba Mahavidyalaya Achalpur, Amravati, Maharashtra, India.
2 Department of Zoology, Shri Shivaji Science College, Amravati, Maharashtra, India.
Corresponding author email: amol_rawankar@yahoo.co.in
Article Publishing History
Received: 06/12/2020
Accepted After Revision: 29/03/2021
DNA Barcoding is considering a novel tool for the identification of species and for discovering the new species by using molecular methods. The mitochondrial cytochrome C oxidase subunit I (COI) serve as fast and accurate method for species identification by molecular methods. The present study was carried out on the three state highways of Vidharbha region, passing through Amravati District which includes Amravati – Chandur Railway state highway (passing through Pohra – Malkhed reserve forest, Amravati- Paratwada state highway passing through agricultural landscape and Paratwada – Semadoh state highway passing through Melghat Tiger reserve. The pectorial muscle tissue samples of the birds killed due to vehicle collision were collected during the study period from 2015 to 2017. Samples on collection immediately dipped in 95 % ethanol, labelled it and brought to the laboratory where they were stored at -20°C for DNA isolation and barcoding analysis.
The generated DNA barcodes has been submitted to gene bank and accession numbers for were received. In this study the DNA barcodes has been generated for 17 road killed avian species. The present study, probably for the first time is reporting the barcode sequences of 6 different birds species from India, as there were no other sequences for these bird’s species found in NCBI database from India. The 6 bird species are Prinia inornata (Plain Prinia), Prinia socialis (Ashy Prinia), Streptopelia senegalensis (Laughing Dove), Otus bakkamoena (Indian Scops Owl), Coturnix backgammon (Common Quail) and Caprimulgus indicus (Indian Nightjar). The present study also demonstrated that the DNA barcoding technique can be accurately applied to the identification of road-killed avian carcasses.
DNA Barcoding, Birds, Road Kills, Vidharbha, India.
Rawankar A. S, Wagh G. A. DNA Barcoding of Some Avian Species Killed due to Road Vehicle Collision in Vidharbha region, India.. Biosc.Biotech.Res.Comm. 2021;14(1).
Rawankar A. S, Wagh G. A. DNA Barcoding of Some Avian Species Killed due to Road Vehicle Collision in Vidharbha region, India.. Biosc.Biotech.Res.Comm. 2021;14(1). Available from: <a href=”https://bit.ly/3kMRkMs“>https://bit.ly/3kMRkMs</a>
Copyright © Rawankar and Wagh This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
In Vidarbha, a total of 417 bird species has been reported including 394 bird species of Amravati District, of which the Melghat Tiger Reserve in Amravati District shows the greatest diversity of avian fauna with recorded 265 bird species (WECS 2009; Wadatkar et al., 2016). DNA Barcoding is considering a novel tool for the identification of species and for discovering the new species by using molecular methods. The mitochondrial cytochrome C oxidase subunit I (COI) serve as fast and accurate method for species identification by molecular methods. Mitochondrial DNA (mt DNA) used extensively in phylogenetic studies of animals as it evolves much more rapidly than nuclear DNA results in accumulation of differences between closely related species (Brown et al., 1979; Savolainen et al., 2005; Hebert et al., 2004, 2010).
Many researchers have suggested that the 648 bp region of mitochondria for mitochondrial cytochrome C oxidase subunit I (COI) serves as the most accurate molecular marker for species identification in animals due to its high interspecific variation, low intraspecific variation, and relatively universal primers for taxonomic groups at the level of orders and even classes (Hebert et al., 2004; Ward et al., 2005; Hajibabaei et al., 2006; Johnsen et al., 2010; Wadatkar et al., 2016). Considerable amount of work and a large number of publications in this field has lead to the formation of Consortium for the Barcode of Life (CBOL, http://barcoding.si.edu), with the objective of obtaining DNA barcodes from all species on the planet (Yoo et al., 2006). By using this approach various animal groups have been studied such as Neotropical bats, North American birds, New Zealand birds, Australian fishes, and tropical Lepidoptera (Ward et al., 2005; Hajibabaei et al., 2006; Clare et al., 2007; Kerr et al., 2007; Tizard et al., 2019).
Species identification through DNA barcoding has many practical utilities such as in conservation biology, food security control, and bird strike identification (Neigel et al., 2007; Dove, 2008; Ward et al., 2008; Wong and Hanner, 2008). Tizard et al. (2019) studied the COI sequence data of New Zealand birds and found that DNA barcoding accurately identified most New Zealand bird species. DNA barcoding provides the rapid method for screening the biodiversity of the particular region for identification of species and also serves as a promising tool for differentiation of two species which have similar or identical phenotypes. It could also be a novel tool for identification of small and immature organisms which are difficult to identify morphologically (Tizard et al., 2019). Birds are the most studied and taxonomically variable class of animals and hence it is very much useful for testing the efficacy of DNA barcoding in species identification. Birds are routinely killed in the road accident by various vehicles in the forest roadside and sometimes it is very difficult to identify the species morphologically hence attempt has been made to develop the DNA barcode for the road killed avian species of Amravati district of Vidharbha region, (Tizard et al., 2019).
MATERIAL AND METHODS
The pectorial muscle tissue samples of the birds killed due to vehicle collision were collected during the study period from 2015 to 2017. The tissues were collected during road vehicle collision study of avian fauna, on the three roads of Amravati District viz., 1. Road passing through reserve forest (Pohra – Malkhed) from Amravati to Chandur Railway (State Highway number -243; 30 Km), 2. Road passing through the agricultural landscape (from Amravati to Paratwada) (State Highway Number -6; 50 km), 3.A road from Paratwada to Semadoh passing through Melghat Tiger reserve (State Highway number -6; 46 Km). Fig No. 1 shows the study area and all the three studies highways.
Figure 1: Map of Amravati District (Study Area)
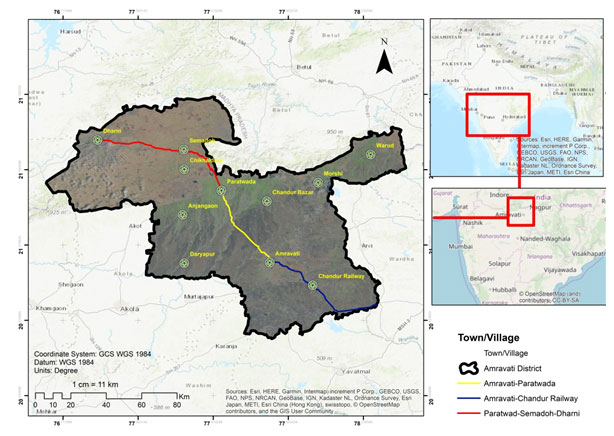
(Sources: Esri, HERE, Garmin, Intermap, increment P Corp., GEBCO, USGS, FAO, NPS, NRCAN, GeoBase, IGN, Kadaster NL, Ordnance Survey, Esri Japan, METI, Esri China (Hong Kong), swisstopo, ©OpenStreetMap contributors, and the GIS User Community; Map Prepared by Shubham Wagh)
Samples on collection immediately dipped in 95 % ethanol, labelled it and brought to the laboratory where they were stored at -20°C for DNA isolation and barcoding analysis. These samples were used for COI gene sequence analysis. Feather sample was used to generate DNA barcode sequence for Indian Peafowl. The feather sample was collected from the Agricultural land. The process of DNA isolation, Polymerase chain reaction, purification of Polymerase chain reaction product and DNA sequencing were carried out at GenOmBiotechnologies Pvt Ltd. Pune. The blast analysis and phylogenetic analysis were carried out in the research laboratory of the Department of Zoology, Shri Shivaji Science College Amravati. DNA was isolated using QIAGEN kit, QIA amp DNA FFPE Tissue (CAT.NO. 56404) as per manufacturer’s instructions.
DNA was eluted in 20.0 μl of elution buffer. Two types of primers were used for the amplification of COI genes: COI gene specific forward and reverse universal primers (Folmers Primer) (Used for birds which include Greater Coucal, Barn Owl, Indian Scops Owl, Spotted Owlet, Ashy Prinia, Indian Nightjar, Plain Prinia, Indian Roller, Red Wattle Lapwing, Cattle Egret, Asian Koel, Common Tailor Bird, Rufous Treepie).Birds specifics primers – BirdF and BirdR (Used for birds which includes Red Vented Bulbul, Laughing Dove, Indian Peafowl and Common Quail). COI gene specific forward and reverse universal primers were used for then amplification is:LCO- 1490 5’- GGTCAACAAATCATAAAGATATTGG- 3’ and HCO –2198 5’- TAAACTTCAGGGTGACCAAAAAATCA – 3’
PCR reaction mix was prepared for DNA samples. Final volume of each reaction was 25.0 μl. Thermal cycling program for PCR was used as below: Initial denaturation at 95 oC for 5 min, followed by 35 cycles of denaturation at 94o C for 1 min, annealing at 40 o C for 1 min, Extension at 72 OC for 1.30 min and final extension at 72 o C for 7 min and hold at 4 o c until use.
Birds specifics primers – BirdF and BirdR: COI gene specific forward and reverse primers were used for the amplification are:
| Primer Name | Primer Sequences |
| BirdF | 5’-TTCTCCAACCACAAAGACATTGGCAC-3’ |
| BirdR | 5’-ACGTGGGAGATAATTCCAAATCCTG-3’ |
PCR reaction mix was prepared for DNA samples. Final volume of each reaction was 25.0 μl. Thermal cycling program for PCR was used as below: Initial denaturation at 95 oC for 5 min, followed by 40 cycles of denaturation at 94o C for 0.45 Sec, annealing at 58 o C for 0.45 Sec, Extension at 72 OC for 1.00 min and final extension at 72 o C for 7 min and hold at 4 o c until use. Agarose gel electrophoresis of the PCR products was performed using 2% (w/v) agarose gel using standard 0.5X TBE gel electrophoresis buffer. Sizes of the amplicons generated by this primer pair is 708 bp and is also evident from the gel. The purification of all amplicons was performed using Purelink PCR product purification kit from Life technologies as per the manufacturer’s instructions. The purified PCR products were again checked on 2% Agarose gel to confirm that the amplicons are not lost during purification.
DNA sequencing of PCR product was performed using both the primers using Applied Biosystems BigDye Terminator V3.1 Cycle sequencing kit. The sequencing products were loaded on Applied Biosystems 3130 Genetic Analyzer – automated DNA sequencing instrument. Sequences were analyzed using Sequencing Analysis 5.1 software available in the sequencing machine. These sequences were further copied and analyzed using ChromasPro v 1.34. Forward and reverse sequences were aligned to form the contig with best sequence calls, hence for each sample using two sequences (Forward and Reverse), one contig was generated. Contig sequence was further subjected to BLAST analysis using http://blast.ncbi.nlm.nih.gov/Blast.cgi tool available at
RESULTS AND DISCUSSION
In the present study, the DNA barcoding of 17 bird species, which were killed due to road vehicle collision has been done. The DNA Barcode sequence of all the 17 species was submitted to the GenBank and the accession numbers were collected. The present study demonstrated that the DNA barcoding technique can be accurately applied to the identification of road-killed avian carcasses. DNA barcoding method was found significant for the identification of road-killed birds’ species.
Table 1. The species for which DNA Barcodes have been generated along with their GenBank Accession Number
| Sr. No | Common Name | Scientific Name | Accession No. |
| 1 | Indian Nightjar | Caprimulgus indicus | KT240050 |
| 2 | Common Tailor Bird | Orthotomus sutorius | KT240051 |
| 3 | Plain Prinia | Prinia inornata | KT240052 |
| 4 | Indian Roller | Coracias benghalensis | KT240053 |
| 5 | Red Wattle Lapwing | Vanellus indicus | KT240054 |
| 6 | Cattle Egret | Bubulcus ibis | KT240055 |
| 7 | Asian Koel | Eudynamys scolopaceus | KT240056 |
| 8 | Greater Coucal | Centropus sinensis | KT240057 |
| 9 | Ashy Prinia | Prinia socialis | KT240058 |
| 10 | Rufous Treepie | Dendrocitta vagabunda | KT240059 |
| 11 | Red Vented Bulbul | Pycnonotus cafer | KT240060 |
| 12 | Laughing Dove | Streptopelia senegalensis | KT240061 |
| 13 | Barn Owl | Tyto alba | KR779892 |
| 14 | Indian Scops Owl | Otus bakkamoena | KR779893 |
| 15 | Spotted Owlet | Athene brama | KR779894 |
| 16 | Indian Peacock | Pavo cristatus | MN206039 |
| 17 | Common Quail.
|
Coturnix cristates | MN206040 |
The present study, probably for the first time has reported the barcode sequences of 6 different bird species from India, as there were no other sequences for these bird species found in NCBI database from India. The 6 birds’ species are Prinia inornata (Plain Prinia), Prinia socialis (Ashy Prinia), Streptopelia senegalensis (Laughing Dove), Otus bakkamoena (Indian Scops Owl), Coturnix cristates (Common Quail) and Caprimulgus indicus (Indian Nightjar). The species for which DNA Barcodes have been generated along with their GenBank Accession Number are shown in table No.1.
Large number of bird species have been studied through DNA barcoding worldwide, such as DNA Barcoding of Korean Birds, Scandinavian birds, critically endangered bird species Asian Houbara Bustard, Netherlands birds, feather mite studies, to identify bird species involved in bird-aircraft collision (Yoo et al., 2006; Dove, 2008; Johnsen et al., 2010; Arif et al., 2012; Aliabadian et al., 2013; Dona et al., 2015; Gaikwad et al., 2016). Tizard et al.
(2019) studied DNA Barcoding in New Zealand birds and demonstrates that DNA barcoding can identify the majority of New Zealand birds to the species level. Many of the times birds are get killed by the road vehicle collision. Goncalves et al. (2015) report a case in which DNA Barcoding technique help criminal investigations and to design species-specific anti-poaching strategies, and also demonstrate how DNA sequence analysis in the identification of bird species is a powerful conservation tool (Goncalves et al., 2015; Tizard et al., 2019).
Dimitriou et al. (2017) studied DNA barcoding of the large majority of bird species resident in Cyprus plus several migrants that were illegally captured. Their study was carried out to support local authorities in their anti-poaching actions. Gaikwad et al. (2016) studied the utility of DNA barcoding for the identification of bird-strike samples from India. They have evaluated the utility of DNA barcoding for species identification of birds involved in bird-strike incidences and concluded that DNA barcoding offers a fast and reliable technique for species identification compared to traditional methods because if blood spots or damaged tissues are provided, traditional methods very often failed to identify the species. In agreement with the above studies, the present study also demonstrated that the DNA barcoding technique can be accurately applied to the identification of road-killed avian carcasses (Gaikwad et al., 2016; Dimitriou et al., 2017).
DNA barcoding method was found significant for the identification of road-killed birds’ species (Gaikwad et al., 2016; Dimitriou et al., 2017). As mentioned above, the DNA Barcoding offers a range of applications in various situations to identify the species. Birds are routinely killed in the road accident by various vehicles in the forest roadside and sometimes it is very difficult to identify the species morphologically due to complete pressing of the animal, hence attempt has been made to develop the DNA barcode for the road killed avian species of Amravati district (Dimitriou et al., 2017).
CONCLUSION
In this study the DNA barcodes has been generated for 17 road vehicle collision killed avian species. The study uses the road killed bird specimens for tissue collection. Total 17 bird’s DNA Barcode sequences were generated and submitted to Genbank. The present study, probably for the first time reporting the barcode sequences of 6 different birds’ species from India, as there were no other sequences for these bird’s species found in NCBI database from India. The present study also demonstrated that the DNA barcoding technique can be accurately applied to the identification of road-killed avian carcasses.
ACKNOWLEDGEMENTS
We are thankful to Chief Conservator of Forest and Field Director, Melghat Tiger Reserve, Amravati for providing me the permission for carrying out my research work on the roads passing through Melghat tiger reserve (Letter No. Desk -3/MTR/Res/831/2015-16. Amravati Dated – 14/08/2015). The author (ASR) is grateful to the Council of Scientific and Industrial Research (CSIR) for provided the research fellowship for carrying out his research work. We are thankful to Dr. J. S. Wadatkar, Mr. Jagdev Iwane, Mr. Hayat Qureshi, Mr. Prathmesh Tiwari and all other Wildlife and Environment Conservation Society (WECS) volunteers for their invaluable help and support. We are also thankful to Mr. Shubham Wagh for preparing the Map of Amravati District showing the study area.
Authors Contributions: Both the authors have equal contribution in bringing out this research work.
Conflict of Interest: The authors declare that they have no Conflict of interests.
REFERENCES
Aliabadian, M., Beentjes, K.K., Roselaar, C.S., Van Brandwijk H., Nijman, V., and Vonk, R. (2013). DNA barcoding of Dutch birds. ZooKeys, 365: 25–48.
Arif, I., Khan H., Williams, J. Shobrak, M. and Arif W. (2012). DNA Barcodes of Asian Houbara Bustard (Chlamydotis undulata macqueenii). International Journal Molecular Sciences 13: 2425-2438
Brown, W. M., George, M. Jr., and Wilson, A. C. (1979). Rapid evolution of animal mitochondrial DNA. Proceedings of National Academy of Science, U.S.A.76:1967-1971.
Clare, E.L., Lim, B.K., Engstrom, M.D., Eger, J.L., and Hebert, P.D.N. (2007). DNA barcoding of Neotropical bats: species identification and discovery within Guyana. Molecular Ecology Notes. 7:184–190.
Dimitriou, A.C., Forcina, G., Papazoglou, C., Panayides, P., Guerrini, M., Crabtree, A., Barbanera, F. and Sfenthourakis, S. (2017). DNA barcoding of bird species in Cyprus: a tool for conservation purposes. Bird Conservation International, page 1 of 12. BirdLife International, 2017 doi:10.1017/S0959270916000472
Dona, J., Diaz- Real, J., Mironov, S., Bazaga, P., Serrano, D., and Jovani, R. (2015). DNA barcoding and mini-barcoding as a powerful tool for feather mite studies. Mol Ecol Resour. doi: 10.1111/1755-0998.12384
Dove, C.J., Rotzel, N.C., Heacker, M., and Weigt, L.A. (2008). Using DNA barcodes to identify bird species involved in birdstrikes. Journal of Wildlife Management.72:1231–1236.
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994) DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol, 3, 294- 299.
Gaikwad, S.P., Munot, H., and Shouche, Y. S. (2016). Utility of DNA barcoding for identification of bird-strike samples from India. Current Science, 110 (1): 25 – 28
Gonçalves, P.F.M., Adriana, RO-M, Tania, E.M., and Cristina, Y.M. (2015). DNA Barcoding Identifies Illegal Parrot Trade. Journal of Heredity, 560–564
Hajibabaei, M., Janzen, D.H., Burns, J.M., Hallwachs, W., and Hebert, P.D.N. (2006). DNA barcodes distinguish species of tropical Lepidoptera. Proc Proceedings of the National Academy of Sciences. 103:968–971.
Hebert, P.D.N., de Waard, J.R., and Landry, J.F. (2010). DNA barcodes for 1/1000 of the animal kingdom. Biology Letters. 6: 359–362.
Hebert, P.D.N., Stoeckle, M.Y., Zemlak, T.S., and Francis, C.M. (2004). Identification of birds through DNA barcodes. PLoS Biology. 2(10): 312.
Johnsen, A., Rindal, E., Ericson, P.G.P., Zuccon, D., Kerr, K.C.R., Stoeckle, M.Y., and Lifjeld, D. (2010). Fish species. Philosophical Transactions of the Royal Society B. 360: 1847–1857.
Kerr, K.C.R, Stoeckle, M.Y., Dove, C.J., Weigt, L.A., Francis, C.M., and Hebert, P.D.N. (2007). ComprehensiveDNA barcode coverage of North American birds. Molecular Ecology Notes. 7:535–543.
Neigel, J., Domingo, A., and Stake, J. (2007). DNA barcoding as a tool for coral reef conservation. Coral Reefs. 26: 487–499.
Savolainen, V., Cowan, R.S., Vogler, A.P., Roderick, G.K., and Lane, R. (2005). Towards writing the encyclopedia of life: an introduction to DNA barcoding. Philosophical Transactions of the Royal Society B. 360: 1805–1811.
Tizard, J., Patel, S., Waugh, J., Tavares, E., Bergmann, T., Gill, B., Norman, J., Christidis, L., Scofield, P., Haddrath, O., Baker,A., Lambert, D., and Miller, C. (2019). DNA barcoding a unique avifauna: An important tool for evolution, systematics and conservation. BMC Evol. Biol. 19: 52
Wadatkar, J., Kasambe, R., and Wagh, G. A. (2016). Updated Checklist of Birds of Amravati district.
Ward, R.D., Holmes, B.H., White, W.T., and Last, P.R. (2008). DNA barcoding Australasian chondrichthyans: results and potential uses in conservation. Marine and Freshwater Research. 59:57–71.
Ward, R.D., Zemlak, T.S., Innes, B.H., Last, P.R., and Hebert, P.D.N. (2005). DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B.360:1847–1857.
WECS (2009). A checklist of the birds of Vidarbha. Wildlife and Environmental Conservation Society Amravati. pp 30.
Wong, E.H.K., and Hanner, R.H. (2008). DNA barcoding detects market substitution in North American seafood. Food Research International .41:828– 837
Yoo, H.S., Eah, J.Y., Kim, J.S., Kim, Y.J., Min, M.S., Paek, W.K., Lee, H., and Kim, C.B. (2006). DNA barcoding Korean birds. Molecules and Cells, 22: 323–327.


