1Ho Chi Minh City University of Food Industry, Ho Chi Minh City, Vietnam.
2Sub-Department of Cultivation, Plant Protection and Quality Control of
Agricultural Products, Long An Provovince, Vietnam.
Corresponding author email: thehv@hufi.edu.vn
Article Publishing History
Received: 17/06/2021
Accepted After Revision: 20/09/2021
Jackfruit (Artocarpus heterophyllus) is one of the popular fruit trees grown in tropical countries. This tree is grown popularly since it has many uses in cuisine, aesthetics as well as in medicine. Although Vietnam is located in an area suitable for the growth of jackfruit trees, the cultivation of this fruit tree is mainly spontaneous on a small scale and the recent studies carried out on jackfruit mostly focused on the evaluation of agronomical criteria, describing the botanical characteristics and using traditional breeding techniques that are low accuracy leading to low quality of jackfruit, uneven fruit, and low economic efficiency. Currently, DNA barcoding techniques are a highly reliable method for genetic diversity assessment and plant taxonomy, in which the matK and rbcL regions are commonly used in the classification of different crops.
In this study, matK and rbcL loci from jackfruit accessions of Vietnam were sequenced and searched for homology in NCBI Genbank to identify Latin names. The obtained sequences from Vietnam jackfruit were then combined with published corresponding sequences of jackfruit from different countries and targeted for alignment and phylogenetic analysis. The obtained data present the significant variation in both examined DNA barcode loci among jackfruit accessions. The sequence alignment also reveals the distinct variation in rbcL regions of jackfruit accessions in Vietnam in the comparison to rbcL sequences of jackfruit accession from other countries. The obtained results show high effectiveness of using rbcL in classifying jackfruit. Furthermore, it could be helpful tool for scientists in managing, conserving and developing genetic resources for breeding programs, it also has the potential to promote the building of an authentication process of jackfruit in Vietnam.
DNA Barcode, Jackfruit, Matk, Rbcl.
Ngo T. K. A, Nguyen M. P, Tran T. A. T, Tran T. M. T, Ho V. T. Comparing the Effectiveness of MatK and rbcL Barcode Loci to Authenticate Jackfruit Artocarpus heterophyllus Growth in Vietnam. Biosc.Biotech.Res.Comm. 2021;14(3).
Ngo T. K. A, Nguyen M. P, Tran T. A. T, Tran T. M. T, Ho V. T. Comparing the Effectiveness of MatK and rbcL Barcode Loci to Authenticate
Jackfruit Artocarpus heterophyllus Growth in Vietnam. Biosc.Biotech.Res.Comm. 2021;14(3). Available from: <a href=”https://bit.ly/3mb2cqi“>https://bit.ly/3mb2cqi</a>
Copyright © Ngo et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Jackfruit (Artocarpus heterophyllus) is a perennial tree commonly found in several countries such as India, Bangladesh, Myanmar, Sri Lanka, South China, Nepal, Laos, Vietnam, Cambodia, Malaysia, Philippines, Indonesia and throughout Africa, Australia. Presently, Bangladesh, India, Myanmar, Thailand, Vietnam, China, the Philippines, Indonesia, Malaysia and Sri Lanka are major jackfruit producers (Sidhu 2012). Jackfruit is widely grown since it has many uses from cuisine, aesthetics, medicine as well as in construction. Jackfruit wood is hard, durable and easy to saw or carve. It is used to build homes, manufacture high-quality furniture, and make musical instruments such as violins. Jackfruit contain high nutrition contents such as carbohydrates, proteins, vitamins, minerals and phytochemical so it is considered as importance food and feed. Jackfruit seeds can be prepared in many ways such as boiled, roasted or soaked in syrup. The leaves and pods of the fruit are an excellent source of food for livestock such as cows, goats, and sheep (Ranasinghe et al. 2019; Srivastava and Singh 2020).
Mature jackfruit is used in salads or used as a vegetable. Ripe jackfruit can be used as a dessert. In addition, pureed jackfruit is also produced into baby food, juice, jam, and jelly. Freeze-drying, vacuum frying and freezing are new preservation methods for modern jackfruit products (Swami et al. 2012). Furthermore, jackfruit is also traditionally applied to treat several diseases such as asthma, ulcers, wound healing, cough, hypertension (Srivastava and Singh 2020). Usually, jackfruit species are distinguished from each other by the stems, leaves and flowers on the mature tree (Chandrashekar et al.
2018; Dey and Baruah 2019; Dhakar et al. 2020). However, this method is often affected by external factors, the stage of tree development or lack of sufficient morphological variations. The morphological characteristics of many types of jackfruit are very similar in the non-flowering stage. However, when the plants are young, the morphological characteristics have many similarities leading to difficulties in distinguishing varieties (Bogale et al. 2020).
In addition, jackfruit is a cross-pollination plant, seed-based propagation will not guarantee the genetic characteristics as well as the desired characteristics in the offspring compared to the mother plant, so the common method is through asexual propagation. However, if the first parent plant is not correctly identified, all subsequent seedlings will be affected (Sherif et al. 2020). This puts the importance of accurately identifying plants at the molecular level to ensure seedling quality.
Recently, molecular markers have been applied to identify plant and animal varieties that are accurate to the molecular level and are not affected by environmental influences and the developmental stage of the organism. Today, molecular markers are increasingly applied for classifying and analyzing the genetic diversity and characterization of plant species. Compared with traditional markers, molecular markers are easier to perform under laboratory conditions, giving fast results and high accuracy (Chesnokov et al. 2020; Hariharan and Prasannath 2021).
Therefore, molecular markers become effective support tools allowing accurate assessment of the genetic diversity of medicinal plants for conservation, selection and correct identification of plant species, serving for further studies on breeding and breeding of important fruit tree species (Kreuzer et al. 2019; Wu et al. 2019). DNA barcoding is one of molecular markers developed recently, it has begun to be applied intensively in plants. In (2009) the International DNA Barcoding Organization (CBOL) recommended the use of DNA barcoding from the rbcL and matK genes in plant research.
This method is presently used to serve the classification, biodiversity assessment and genetic resource conservation. Several gene regions have been utilized as barcodes for plant classification such as ITS, matK, rbcL, atpF-atpH, psbK-psbI and trnH-psbA. Among them, matK and rbcL have been chosen as standard plant barcoding loci by The Consortium for the Barcode of Life (CBOL Plant Working Group 2009; Swami et al. 2012).
These two loci have been sucessfully applied to classified several plant species such as Cymodocea seagrass genus, jewel orchid, Pseuderanthemum palatiferum (Bchir et al. 2019; Ho and Bui 2021; Ho et al. 2021). The aim of present study was to investigate the discrimination ability in classification of jackfruit accessions collected in Vietnam and from other countries based one of DNA sequences of matK and rbcL loci. The archived results in this study would be applicable for authentication, genetic conservation and breeding purposes of jackfruit in Vietnam.
MATERIAL AND METHODS
DNA from jackfruit leaves was extracted by Cetyltrimethyl Ammonium Bromide (CTAB) method followed Allen et al (Allen et al. 2006). After extraction, DNA quality was examined by electrophoresis on 1% agarose then spectrophotometer (Optima SP 3000 nano UV-VIS, Japan) was used to determine DNA concentrations. The DNA samples were then kept at -20 °C freezer until use for PCR reactions.
MatK and rbcL regions were amplified using the PCR method with composition reactions as follows: 12.5 μL 2X Mytaq Red Mix (Bioline, UK), 20 ng DNA, 0.2 μM of each primer (either matK 390F: 5’-CGATCTATTCATTCAATATTTC-3’; and 1326R: 5’- TCTAGCACACGAAAGTCGAAGT-3’ or rbcL: cF: 5’- TGAAAACGTGAATTCCCAACCGTTTATGCG-3’; cR: 5’- GCAGCAGCTAGTTCCGGGCTCCA-3’ and PCR water for a final volume of 25 μl (Hasebe et al. 1994; Cuénoud et al. 2002).
The PCR cocktails were run in thermal cyclers SureCycler 8800 Thermal Cycler (Agilent, USA) with following conditions: initial denaturation at 94 °C for 2 minutes; then repeated by 35 cycles of 30 seconds at 94 °C, 30 seconds at 55 °C, 50 seconds at 72 °C, and finally one minute at 72 °C to complete the reaction. The PCR products were then stained with 6X GelRed (Biotum, UK) and visualized on gel electrophoresed and 1 kb ladder (Bioline, UK) was used to determine amplification length. Correct PCR products were sequenced by Sanger methods at Nam Khoa Company (Ho Chi Minh City, Vietnam).
Each sample were sequenced for both sense and antisense directions. Antisense sequences were reversed and aligned with sense sequences to ensure accuracy. Furthermore, the all published sequences of matK and rbcL sequences of Artocarpus heterophyllus available on NCBI Genbank http: //www.ncbi.nlm.nih.gov) were downloaded and evaluated as criteria proposed by Suesatpanit and colleagues (2017) (1) sequences are not ‘unverified’ without a species name (2) contain <3% ambiguous base ‘N’. All sequences were then included for alignment and phylogeny analysis (Suesatpanit et al. 2017).
The DNA sequences from Vietnam jackfruit accessions was checked for homology by using Basic Local Alignment Tools (BLAST) (NCBI, USA) database. These sequences will be combined with NCBI-archived sequences then aligned using the Clustal method in Molecular Evolutionary Genetics Analysis (MEGA) 6 software (https://www.megasoftware.net). Neighbour Joining (NJ) and Maximum Likelihood (ML) phylogeneny analysis were then performed and compared since they represent for distance methods and discrete character methods, respectively. To increase the accuracy in phylogenetic construction, 1000 replicates was applied for bootstrap analysis (Kress et al. 2005).
RESULTS AND DISCUSSION
Sequence retrieve: After matK and rbcL loci were successfully amplified and sequenced, the sequences were targeted to find homology using BLAST. However, both matK and rbcL sequences are not applicable for identification of jackfruit due to the returned BLAST results showing the presence of several species in Artocarpus genus such as Artocarpus hypargyreus, Artocarpus altilis or Artocarpus camansi. This result is in agreement with previous study in Indian when 6 jackfruit varieties by using matK gene and the result showing the high sequence variation among this locus (Vazhacharickal et al. 2017).
By using keyword “species names + matK/rbcL” to find the sequences deposited in NCBI GenBank, after removal of unrealizable sequences as Suesatpanit e al. (2017), total of 27 sequences were obtained, there are 8, and 19 DNA sequences of matK and rbcL regions, respectively (Table 1). In general, rbcL locus was more intensively studied with up to 19 sequences acquired these sequences mostly come from USA, Sri Lanka and Japan. Whereas, matK are mostly from USA, Thailand and Sri Lanka have one accession for each country (Suesatpanit et al. 2017; Ho et al. 2021).
Estimation of sequence divergence: The number of base substitutions per site from averaging over all sequence pairs within each group are shown in Table 1 and Table 2. The variation among matK and rbcL regions is from 0.00 – 0.99, 0 – 0.06, and 0.00 – 1.85, respectively.
Table 1. Estimates of Evolutionary Divergence over Sequence Pairs between Groups for matK
| matK_MH748883.1_USA | 0.08 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| matK_MK264372.1_Sri_Lanka | 0.99 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | |
| matK_KU856361.1_USA | 0.00 | 0.99 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| matK_KU856360.1_USA | 0.01 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| matK_KU856359.1_USA | 0.00 | 0.99 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| matK_KU856358.1_USA | 0.00 | 0.99 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| matK_KU856357.1_USA | 0.01 | 1.02 | 0.01 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | |
| matK_LC461814.1_Thailand | 0.01 | 0.99 | 0.01 | 0.00 | 0.01 | 0.01 | 0.02 | 0.00 | 0.00 | 0.00 | |
| matK_Vietnam_1 | 0.00 | 0.99 | 0.00 | 0.01 | 0.00 | 0.00 | 0.01 | 0.01 | 0.00 | 0.00 | |
| matK_Vietnam_2 | 0.01 | 0.99 | 0.01 | 0.00 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.00 | |
| matK_Vietnam_3 | 0.00 | 0.99 | 0.00 | 0.01 | 0.00 | 0.00 | 0.01 | 0.01 | 0.00 | 0.01 |
(Standard error of comparison is presented in italics upper diagonal).
Table 2. Estimates of Evolutionary Divergence over Sequence Pairs between Groups for rbcL
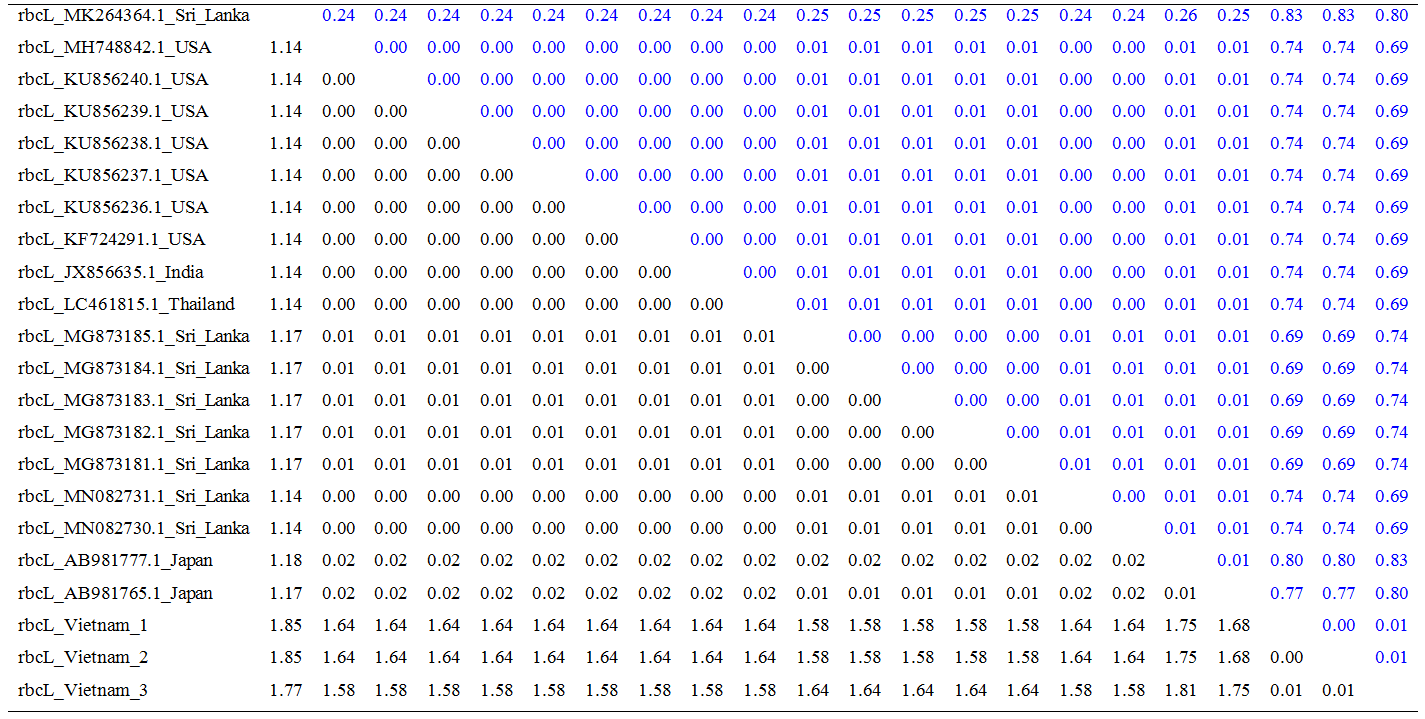
(Standard error of comparison is presented in italics upper diagonal).
Substitution bias consisting of transition and transversion at codon position for each luster could reveal the trend of evolution. In this study, the substitution of different bases in analyzed regions is evaluated on entire codon positions (1st+ 2nd + 3rd nucleotide) and shown in Table 3. In general, transitional substitution is higher than transversional substitution in all loci. rbcL show a higher transversionsal substitution than matK.
Table 3. Maximum Composite Likelihood Estimate of the Pattern of Nucleotide Substitution (in percentage)
| matK | rbcL | |||||||
| A | T | C | G | A | T | C | G | |
| A | – | 12.15 | 5.67 | 1.02 | – | 11.87 | 10.52 | 3.31 |
| T | 9.89 | – | 9.88 | 5.29 | 12.82 | – | 0.12 | 11.09 |
| C | 9.89 | 21.18 | – | 5.29 | 12.82 | 0.14 | – | 11.09 |
| G | 1.9 | 12.15 | 5.67 | – | 3.83 | 11.87 | 10.52 | – |
(Note: Each entry shows the probability of substitution from one base (row) to another base (column). For simplicity, the sum of r values is made equal to 100. Rates of different transitional substitutions are shown in bold and those of transversionsal substitutions are shown in italics).
Estimation of species resolution: Based on phylogenetic analysis, the accession resolution is variable between two DNA barcode loci (Figure 1, and 2). Accession from Sri Lanka is branched into a separate group and remaining accessions are grouped together (Figure 1). Nevertheless, the sequence number should be increased for analysis to enhance the accuracy of study (Sikdar et al. 2018).
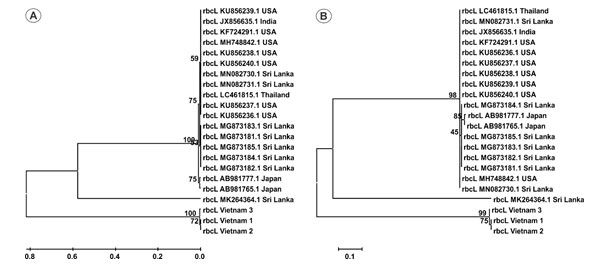
Whereas rbcL locus is able to distinguish jackfruit accessions from Vietnam with those from other countries (Figure 2). The high discrimination power of rbcL found in this study is in line with previous reported as a good marker to differentiate species in several plant species such as suaeda, jewel orchid. After surveying over 10,000 rbcL sequences from Genbank, Newmaster and colleagues also reported that this region is effective for plant classification (Newmaster et al. 2006; Ho et al. 2021).
Figure 1: Phylogenetic tree of 11 matK sequences by UPGMA (A) and Maximum- Likelihood (B) methods with 1000 bootstrap replicates.
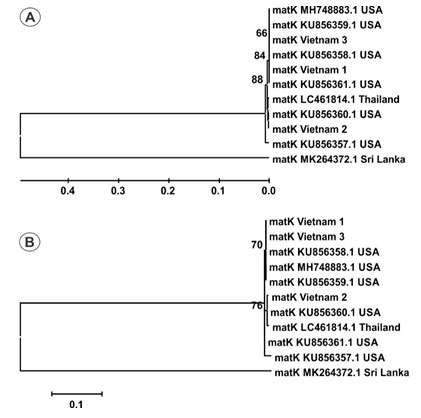
Figure 2: Phylogenetic tree of 22 rbcL sequences by UPGMA (A) and Maximum- Likelihood (B) methods with 1000 bootstrap replicates.
CONCLUSION
The findings of the present study suggest that the discrimination capacity of matK and rbcL DNA barcode loci are variable, in which rbcL locus reveals more potential for classification of jackfruit. In the future, higher sequence number should be included in the analysis to give more reliable result. The information from this study could be useful in conservation and development programs of jackfruit plants. The usefulness of two main DNA barcode loci in classify different jackfruit accessions was investigated.
ACKNOWLEDGEMENTS
This study was supported by The Youth Incubator for Science and Technology Program, managed by Youth Development Science and Technology Center – Ho Chi Minh Communist Youth Union and Department of Science and Technology of Ho Chi Minh City, the contract number is “22/2020/HD-KHCNT-VU”. Moreover, authors would like to thank Ho Chi Minh University of Food Industry for providing research fund and laboratory facility.
Conflict of Interests: Authors declare no conflicts of interests to disclose.
REFERENCES
Allen GC, Flores-Vergara MA, Krasynanski S, et al. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nature Protocols Vol 1 No 5 pages 2320–2325. https://doi.org/10.1038/nprot.2006.384.
Sidhu AS (2012). Jackfruit Improvement in the Asia-Pacific Region: A Status Report. APAARI pages 182.
Bchir R, Djellouli AS, Zitouna N, et al. (2019). Morphology and genetic studies of Cymodocea seagrass genus in Tunisian coasts. Phyton Vol 88No 2 Pages 171-184.
Bogale M, Baniya A and Digennaro P (2020). Nematode Identification Techniques and Recent Advances. Plants Vol 9, 1260 doi:10.3390/plants9101260.
CBOL Plant Working Group (2009). A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America Vol 106 No 31 Pages 12794–12797. https://doi.org/10.1073/pnas.0905845106.
Chandrashekar KG, Vijayakumar RM, Subramanian S, et al. (2018). Morphological Characterization of Jackfruit (Artocarpus hetrophyllus Lam.) Local Genotypes under Coffee Ecosystem of Lower Pulney Hills. International Journal of Current Microbiology and Applied Sciences Vol 7 No 3 Pages 2210-2224.
Chesnokov YV, Kosolapov VM, Savchenko IV (2020). Morphological genetic markers in plants. Russian Journal of Genetic Vol 56 Pages 1406-2425.
Cuénoud P, Savolainen V, ChatrouLW, et al. (2002). Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. American Journal of Botany Vol 89 No 1 Pagas 132–144. https://doi.org/10.3732/ajb.89.1.132.
Day B and Baruah K (2019). Morphological Characterization of Jackfruit (Artocarpus heterophyllus Lam.) of Assam, India. International Journal of Current Microbiology and Applied Sciences Vol 8 No 11 page 1005-1016.
Dhakar MK, Das B, Sarkar PK, et al. (2020). Diversity in jackfruit (Artocarpus heterophyllus Lam.): insights into fruit characterization for the identification of superior genotypes. Plant Genetic Resources: Characterization and Utilization; 1–9. doi:10.1017/S1479262120000325.
Hariharan G and Prasannath K (2021). Recent Advances in Molecular Diagnostics of Fungal Plant Pathogens: A Mini Review. Frontiers in Cellular and Infection Microbiology 10:600234. https://doi.org/10.3389/fcimb.2020.600234.
Hasebe M, Omori T, Nakazawa M, et al. (1994). rbcL gene sequences provide evidence for the evolutionary lineages of leptosporangiate ferns. Proceedings of the National Academy of Sciences of the United States of America Vol 91 Pages 5730–5734.
Ho VT and Bui ST (2021). Comparison of three DNA barcode loci for distinguishing Hoan-ngoc (Pseuderanthemum palatiferum) from its relative. Bioscience Research Vol 18 No 2 pages 1277-1283.
Ho VT, Tran TKP, Vu TTT, et al. (2021). Comparison of matK and rbcL DNA barcodes for genetic classification of jewel orchid accessions in Vietnam. Journal of Genetic Engineering and Biotechnology Vol 19 No 93. https://doi.org/https://doi.org/10.1186/s43141-021-00188-1.
Kress WJ, Wurdack KJ, Zimmer EA, et al. (2005). Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences of the United States of America Vol 102 No 23 Pages 8369–8374. https://doi.org/10.1073/pnas.0503123102.
Newmaster SG, Fazekas AJ and Ragupathy S (2006). DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Canadian Journal of Botany Vol 84 No 3 Pages 335–341. https://doi.org/10.1139/b06-047.
Ranasinghe RASN, Maduwanthi SDT and Marapana RAUJ (2019). Nutritional and Health Benefits of Jackfruit (Artocarpus heterophyllus Lam.): A Review. International Journal of Food Science Vol 2019. Volume 2019, Article ID 4327183. https://doi.org/10.1155/2019/4327183.
Sherif NA, Kumar TS and Rao MV (2020). DNA barcoding and genetic fidelity assessment of micropropagated Aenhenrya rotundifolia (Blatt.) C.S. Kumar and F.N. Rasm.: a critically endangered jewel orchid. Physiology and Molecular Biology of Plants Vol 26 Pages 2391–2405.
Sikdar S, Tiwari S, Thakur VV et al. (2018). An in silico approach for evaluation of rbcL and matK loci for DNA barcoding of fabaceae family. International Journal of Chemical Studies Vol 6 No 6 Pages 2446–2451.
Srivastava R and Singh A (2020). Jackfruit (Artocarpus heterophyllus Lam) Biggest Fruit with High Nutritional and Pharmacological Values: A Review. International Journal of Current Microbiology and Applied Sciences Vol 9 No 8 Pages 764-774.
Suesatpanit T, Osathanunkul K, Madesis P et al. (2017). Should DNA sequence be incorporated with other taxonomical data for routine identifying of plant species? BMC Complementary and Alternative Medicine Vol 17 No 1 Pages 437. https://doi.org/10.1186/s12906-017-1937-3.
Swami SB, Thakor NJ, Haldankar PM, et al. (2012). Jackfruit and Its Many Functional Components as Related to Human Health: A Review. Comprehensive Reviews in Food Science and Food Safety Vol 11 No 6 Pages 565–576. https://doi.org/https://doi.org/10.1111/j.1541-4337.2012.00210.x
Vazhacharickal PJ, Sajeshkumar NK, Mathew JJ et al. (2017). Studies on genetic relationships among six varieties of jackfruit in Kerala employing the “matK” gene using PCR technique and RFLP markers. 88 pages, Grin Publishing.
Wu F, Li M, Liao B et al. (2019). DNA barcoding analysis and phylogenetic relation of mangroves in Guangdong province, China. Forest Vol 10 No 56; doi:10.3390/f10010056.


