Department of Microbiology, Marwadi University, Rajkot, Gujarat, India
Corresponding author email: vivek.pattani@gmail.com
Article Publishing History
Received: 21/01/2022
Accepted After Revision: 25/03/2022
The haphazard application of chemical fertilizers and pesticides causes tremendous damage to ecosystems and all biota. One of the most effective ways to tackle the threat is to use biofertilizer. Plant growth promoting bacteria (PGPB) are an important bacterial source for microbial fertilizers that can boost agricultural yields by encouraging plant growth. Bacterial isolates isolated from Saurashtra region, Gujarat, India were analysed for their capability to solubilize inorganic ‘P’ from tri calcium phosphate and production of indole acetic acid (IAA) quantitatively by bacterial. Production of ammonia, siderophore and hydrogen cyanide (HCN) by selected bacteria isolates was analysed. Biochemical characterization of selected bacterial isolates was done using Vitek 2 Compact system. Isolate GFS15C2 showed highest amount of phosphate solubilization, followed by isolate GFS07C1 and GFS01C1. Bacterial isolate GFS15C2 produced highest amount of IAA. All bacterial isolates were able produce ammonia.
Eight bacteria isolates were be to produce HCN. Siderophore was produced by 14 bacterial isolates. In biochemical characterization all the bacterial isolates were able to use D-glucose. Based on biochemical characters clustering of bacteria isolates was done using Paleontological statistics software package for education and data analysis(PAST). Using cluster analysis by euclidean distance method based on biochemical characterization isolates GFS16C2 & SCS12C3 was found to have distinct characters than other isolates. The present study attempts to characterize PGPB which could be harnessed to improve plant growth. Several phosphate solubilizers and IAA producers also showed production of siderophores and HCN which suggests that these organisms do possess biocontrol ability. These PGPB microbial inoculants can be utilized to improve agricultural systems or as an alternate means of environmentally friendly plant disease biocontrol.
Bacteria,HCN, IAA,Phosphate,Vitek
Pattani V. B, Kaneriya J. P, Joshi K, Sanghvi G. V. Characterization of Plant Growth-Promoting Activity of Bacteria Isolated from Forest and Coastal Regions of Saurashtra, Gujarat, India. Biosc.Biotech.Res.Comm. 2022;15(1).
Pattani V. B, Kaneriya J. P, Joshi K, Sanghvi G. V. Characterization of Plant Growth-Promoting Activity of Bacteria Isolated
from Forest and Coastal Regions of Saurashtra, Gujarat, India. Biosc.Biotech.Res.Comm. 2022;15(1). Available from: <a href=”https://bit.ly/35XKX5O“>https://bit.ly/35XKX5O</a>
Copyright © This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY). https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provided the original author and sources are credited.
INTRODUCTION
Application of haphazard use of chemicals fertilizers andpesticide leads to incredible harm to ecosystems and all biota.Alternative to these chemical applications, biofertilizer areone of the best solutions to combat the hazard. These microbes can be used as both plant growth promoting bacteria and as biocontrol agent for suppression of plant pathogenic microbes(Kour et al. 2019; Sharma et al. 2020). The most commonly known plant growth promoting bacteria are Bacillus, Rhizobia, Pseudomonas, Azotobacteretc. PGPB, it has the ability to fix nitrogen fixation to solubilize phosphate, iron chelation by siderophores and phytohormone production whereas, being a biocontrol agent, it has the ability to produce HCN and secondary metabolites to suppress the growth of plant pathogenic microbes(Backer et al. 2018; Swarnalakshmi et al. 2020).
Hence, its multifaceted capability makes it appropriate as biofertilizer and biocontrol agent in agriculture field. PGPB compounds can exhibit a number of features that influence plant growth, either directly or indirectly. Auxin, gibberellin, ethylene, and other plant growth regulators are produced, as are siderophores, HCN and antibiotics, as well as phosphate solubilization(Suresh et al. 2020; Rawat et al. 2020).The synthesis of siderophore for iron sequestration and the auxin IAA, which has been linked to plant growth promotion, notably root initiation and elongation, is one of the most significant processes involved in plant promotion(Prasad et al. 2019; Morales-Cedeno et al. 2021).
Microbial inoculants provide a number of benefits over chemical inoculants(Vassilev et al. 2020).They are environmentally friendly, sound sources of renewable nutrients that are necessary for soil health and life(Basu et al. 2021). They also fight abiotic stressors and have antagonistic action against a variety of crop diseases. Based on their capacity to absorb nutrients from the soil, fix atmospheric nitrogen, induce nutrient solubilization, and function as biocontrol agents, a variety of microbial taxa have been commercially employed as effective biofertilizers(Tabassum et al. 2017;Kenneth et al. 2019; Kumar et al. 2021).Plant growth promoting bacteria were isolated from the Gir forest coastal region of Saurashtra, Gujarat. using the soil samples by screening for nitrogen fixing bacteria and further screening was done for phosphate solubilizing bacteria and production of IAA producing bacteria was also done (Kaneriya et al. 2021).
MATERIAL AND METHODS
Bacterial isolates were analysed for their capability to solubilize inorganic ‘P’ from tri calcium phosphate.The soluble P was determined by using colorimetric chloro-stannous reduced molybdo-phosphoric acid blue method(Dipak and Abhijit 2005).Experiments were set in triplicates and phosphate solubilization and change in the pH of the medium were analysed for ten days.The blue colour produced was measured at 660 nm using UV-1800 UV-VIS Spectrophotometer, Shimadzu. The amount of P2O5 solubilized was expressed in P mg/L of the medium.Isolates which showed multiple properties of nitrogen fixation, phosphate solubilization and IAA production were further analysed for siderophore production, quantitative estimation of phosphate solubilization and IAA.
The production of siderophore with the universal chemical test method for quality detection of siderophore production in fluid crops were tested for bacterial isolates (Schwyn and Neilands 1987). This experiment was carried out using a modified method that was providedby Hu and Xu(Arora and Verma 2017). For inferring the presence of siderophore, the tubes were monitored for colour changes from dark blue to light blue or orange. Uninoculated medium was used as control.Production of IAA was estimated by growing the bacterial isolates in yeast extract-mannitol broth and minimal medium supplemented with 200 µg/ml L-tryptophan, which acts as a precursor for IAA synthesis at 28 ± 1⁰C.
After 48 hours, 5 ml cultures were centrifuged at 10,000 RPM for 15 minutes at 4 ± 1⁰C and the supernatant was collected(Pereira et al. 2019).Experiments were set in triplicatesto analyse the production of IAA. Development of pink colour indicates production of IAA and its optical density was recorded at 530nm using UV-1800 UV-VIS Spectrophotometer, Shimadzu. Concentration of IAA produced was estimated against standard curve of IAA (Hi-media) in the range of 10–100 µg /ml.Selected bacterial isolates were analysed for the production of ammonia in peptone water. Cultures were inoculated in 10 ml peptone water and incubated at 28±2 °C for 48 to 72 hours.
Nessler’s reagent (0.5 ml) was added in each tube.Ammonia production was indicated by the change in colour from brown to yellow(Cappuccino and Welsh 2017).Selectedbacterial isolates were analysed for the production of hydrogen cyanide by the method given by Lorck (1948). Parafilm was used to seal the plates, which were then incubated at 28±2 °C for 4 days. HCN production was indicated by the colour changing from yellow to red(Slama et al. 2019).Biochemical characterization of the selected bacterial isolates was done using Vitek 2 compact, Biomerieux Inc.All the descriptive statistics testswere performed using XLSTAT for Windows (Fahmy 2016). Graphs were created using Microsoft Office Excel 2019. Clusteranalysis based on Euclidean distances was done using PAST software by (Hammer et al. 2001).
RESULTS AND DISCUSSION
The present study reflects the work done to characterize plant growth promoting parameters isolated from forest and coastal regions of Saurashtra, Gujarat and analyse their biochemical parameters. For soil microorganisms, rhizosphere is appropriate endurance specialty because of rich supplement accessibility(Khatoon et al. 2020). Community of life forms living in a specific specialty is explicit and subject to the actual elements and ecological variables(Mony et al. 2020).
Phosphate solubilization is one vital attribute of plant development advancement as microorganisms solubilize insoluble phosphate making it accessible for plants(Joshi et al. 2021).Quantitative analysis of phosphate solubilization: Bacterial isolates GFS01C1, GFS03C1, GFS04C1, GFS05C2, GFS07C1, GFS08C1, GFS10C1, GFS11C1, GFS12C1, GFS13C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2, SCS03C1, SCS04C1, SCS07C3, SCS12C1, SCS12C2, SCS12C3, SCS12C4, SCS12C5 and SCS07C2 were analysed for the phosphate solubilization(Mony et al. 2020).
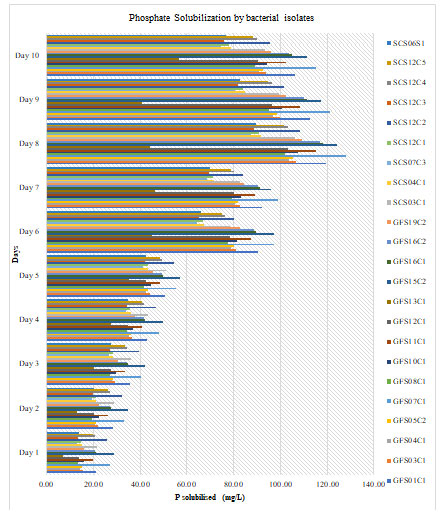
Solubilization of Phosphate by bacterial isolates and pH reduction of the medium for was analysed for 10 days and of which values of P solubilized on day 5 & 10 are tabulated in the Table 1. Graphical representation for P Solubilization by bacterial isolates from day 1 to day 10 has beengiven in figure 1. It was observed that over a period of time, the un-inoculated medium detected no soluble P and no drop in pH as well. In case of inoculated medium solubilization of the phosphate was observed with the decrease in pH. Bacterial isolate GFS15C2 showed highest amount of phosphate solubilization, followed by isolateGFS07C1, GFS01C1, GFS16C1, GFS16C2, GFS11C1, SCS03C1 and SCS12C4. Isolate GFS13C1 showed lowest phosphate solubilization.Marra et al. (2019) reported that all separates which solubilize tri-calcium phosphate (TCP) in fluid and medium could create natural acids causing the pH of the medium acidic.Similarly, all of the isolates that can solubilize TCP in the current investigation demonstrated a decline in the pH of the medium.Similar observation was made Lebrazi et al. (2020). This drop in the pH is supported by secretion of different natural acids by use of sugar(Marra et al. 2019; Lebrazi et al. 2020).
Table 1. Solubilization of Phosphate by bacterial isolates and change in pH on day 5 & 10
(P Indicates Solubilised phosphate and values indicate mean standard error)
| Day | Day 5 | Day 10 | |||||||||||
| Sr. | Isolate | P (mg/L) | pH | P (mg/L) | pH | ||||||||
| 1 | GFS01C1 | 50.48 | ± | 0.12 | 5.11 | ± | 0.05 | 106.33 | ± | 0.68 | 4.19 | ± | 0.11 |
| 2 | GFS03C1 | 44.18 | ± | 1.08 | 5.06 | ± | 0.01 | 93.73 | ± | 2.76 | 4.14 | ± | 0.15 |
| 3 | GFS04C1 | 42.72 | ± | 0.69 | 5.07 | ± | 0.04 | 90.80 | ± | 1.96 | 4.15 | ± | 0.14 |
| 4 | GFS05C2 | 43.53 | ± | 0.94 | 5.10 | ± | 0.02 | 92.42 | ± | 2.18 | 4.18 | ± | 0.16 |
| 5 | GFS07C1 | 55.45 | ± | 0.83 | 5.03 | ± | 0.06 | 115.23 | ± | 1.28 | 4.12 | ± | 0.17 |
| 6 | GFS08C1 | 41.85 | ± | 0.71 | 5.02 | ± | 0.06 | 89.06 | ± | 1.84 | 4.11 | ± | 0.17 |
| 7 | GFS10C1 | 44.51 | ± | 1.38 | 4.97 | ± | 0.07 | 94.38 | ± | 3.37 | 4.06 | ± | 0.20 |
| 8 | GFS11C1 | 48.43 | ± | 0.75 | 5.08 | ± | 0.04 | 102.23 | ± | 2.05 | 4.17 | ± | 0.15 |
| 9 | GFS12C1 | 42.52 | ± | 0.86 | 5.05 | ± | 0.05 | 90.41 | ± | 2.39 | 4.14 | ± | 0.15 |
| 10 | GFS13C1 | 35.15 | ± | 0.36 | 5.41 | ± | 0.15 | 56.75 | ± | 1.01 | 4.50 | ± | 0.19 |
| 11 | GFS15C2 | 57.22 | ± | 1.60 | 4.98 | ± | 0.06 | 111.30 | ± | 2.22 | 4.06 | ± | 0.14 |
| 12 | GFS16C1 | 49.83 | ± | 0.80 | 5.10 | ± | 0.06 | 105.02 | ± | 2.26 | 4.19 | ± | 0.10 |
| 13 | GFS16C2 | 49.20 | ± | 0.79 | 5.06 | ± | 0.04 | 103.76 | ± | 2.24 | 4.15 | ± | 0.14 |
| 14 | GFS19C2 | 45.34 | ± | 1.15 | 5.08 | ± | 0.05 | 96.04 | ± | 2.70 | 4.17 | ± | 0.11 |
| 15 | SCS03C1 | 50.92 | ± | 0.31 | 5.05 | ± | 0.02 | 93.46 | ± | 0.88 | 4.14 | ± | 0.17 |
| 16 | SCS04C1 | 43.53 | ± | 0.64 | 5.10 | ± | 0.04 | 78.68 | ± | 1.50 | 4.19 | ± | 0.12 |
| 17 | SCS07C3 | 41.43 | ± | 0.84 | 5.09 | ± | 0.05 | 74.49 | ± | 2.02 | 4.17 | ± | 0.11 |
| 18 | SCS12C1 | 43.22 | ± | 0.58 | 5.17 | ± | 0.04 | 78.07 | ± | 1.48 | 4.26 | ± | 0.17 |
| 19 | SCS12C2 | 54.41 | ± | 2.00 | 5.07 | ± | 0.06 | 95.51 | ± | 1.38 | 4.15 | ± | 0.20 |
| 20 | SCS12C3 | 42.07 | ± | 0.75 | 5.12 | ± | 0.04 | 75.76 | ± | 1.81 | 4.21 | ± | 0.12 |
| 21 | SCS12C4 | 49.26 | ± | 0.81 | 5.05 | ± | 0.03 | 90.15 | ± | 1.97 | 4.14 | ± | 0.14 |
| 22 | SCS12C5 | 48.43 | ± | 0.75 | 5.14 | ± | 0.05 | 88.49 | ± | 1.85 | 4.22 | ± | 0.20 |
| 23 | SCS06S1 | 42.52 | ± | 0.86 | 5.06 | ± | 0.04 | 76.67 | ± | 2.24 | 4.15 | ± | 0.16 |
Figure 1: Plot for Phosphate Solubilizationby bacterial isolates from day 1 to day 10.
Production of Siderophore: Selected bacterial isolates were analysed for production of siderophoreBacterial isolates GFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2, SCS03C1, SCS07C3, SCS12C1, SCS12C2, SCS12C3, SCS12C5 and SCS06C1 indicated the production of siderophores. Total 14 bacterial isolates produced siderophores.Bacterial isolates were further screened on the basis of siderophore production and were analysed for their capability to produce IAA quantitatively.Other influential traits of PGPB, which can indirectly enhance plant growth is by producing siderophores, chitinase andHCN, which protects plants against pathogens (Athira and Anith 2020; Legein et al. 2020). Under present study, important traits such as siderophore and HCN production have been assessed in vitro.
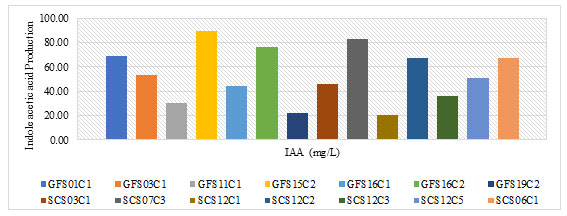
Indole acetic Acid: Bacterial isolatesGFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2,SCS03C1, SCS07C3, SCS12C1, SCS12C2, SCS12C3, SCS12C5 and SCS06C1which indicated the production of siderophores& higher amount of phosphate solubilization were further analysed for the quantitative assay of indole acetic acid. Production of IAA by bacterial isolates analysed is tabulated in the table 2 and plotted in figure 2. Bacterial isolate GFS15C2 (89.43 mg/L) produced highest amount of IAA followed by SCS07C3, GFS16C2, GFS01C1, SCS12C2 and SCS06C1.
Bacterial isolates SCS12C1 lowest.IAA synthesis is an important characteristic of PGPR because this phytohormone allows the plant to form a well-organized root system, which improves nutrient absorption(Kumar et al. 2019). IAA have been reported to stimulate the growth of roots and leaves in plants thereby alleviating plant productivity (Bhat et al. 2020).Among the all the bacterial isolates bacterial isolate GFS15C2 was found produced highest IAA. The IAA produced by bacteria varies between different species and strains. This variation is possibly affected by several factors, such as physiological condition of bacterial isolates, growth stage, and substrate availability(Lebrazi et al. 2020a).
Table 2. Production of IAA by bacterial isolates. (Value indicates mean standard error)
| Isolate | Indole acetic acid (mg/L) | Isolates | Indole acetic acid (mg/L) | ||||
| GFS01C1 | 69.02 | ± | 0.72 | SCS03C1 | 45.87 | ± | 0.93 |
| GFS03C1 | 52.89 | ± | 1.04 | SCS07C3 | 82.77 | ± | 1.02 |
| GFS11C1 | 29.95 | ± | 0.41 | SCS12C1 | 19.97 | ± | 0.89 |
| GFS15C2 | 89.43 | ± | 1.01 | SCS12C2 | 67.07 | ± | 0.74 |
| GFS16C1 | 44.50 | ± | 0.81 | SCS12C3 | 35.81 | ± | 0.54 |
| GFS16C2 | 76.62 | ± | 0.89 | SCS12C5 | 50.65 | ± | 1.01 |
| GFS19C2 | 22.07 | ± | 0.93 | SCS06C1 | 66.93 | ± | 0.89 |
Figure 2: Plot for Production of Indole acetic acid by bacterial isolates.
Ammonia Production: Bacterial isolatesGFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2,SCS03C1, SCS07C3, SCS12C1, SCS12C2, SCS12C3, SCS12C5 and SCS06C1were analysed for the production of ammonia. All isolates were able to produce ammonia.
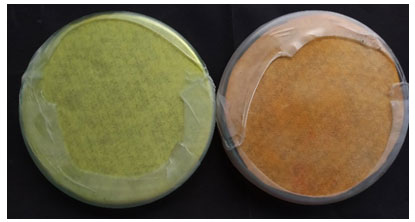
HCN production:Bacterial isolates GFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2, SCS03C1, SCS07C3, SCS12C1, SCS12C2, SCS12C3, SCS12C5 and SCS06C1were analysed for the production of HCN.Week HCN production was indicated by a little change in the colour of Whatman filter paper, with yellow-coloured paper turning pale orange. Bacterial isolates GFS01C1, GFS11C1, GFS15C2, GFS16C2, GFS19C2, SCS03C1, SCS07C3 and SCS12C2 were able to produce HCN.
Figure 3: HCN production by bacterial isolates yellow coloured plate (negative control) and orange coloured which indicated the production of HCN.
Total 14 isolates showed siderophore production and 8 showed HCN production. Several of the strains that could inhibit pathogenic Fusarium oxysporum growth also had the potential to produce HCN, siderophores, and antibiotics(Karthika et al. 2020, Bandopadhyay et al. 2019). As a result, indirect PGP features are just as significant as direct PGP factors for total plant development(Shastri et al. 2020).
Biochemical characterization of bacterial isolatesGFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2, SCS03C1, SCS07C3, SCS12C1, SCS12C2, SCS12C3, SCS12C5 and SCS06C1 was done using Vitek 2 compact and data wastabulated in the table 3& 4.Byuse of Vitek 2 compact, 47 biochemical characterization tests such as carbohydrate fermentation, lipase, decarboxylases, phosphatases etc can be analysed. All the isolates where able to ferment the D-glucose. Bacterial isolates GFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C2, GFS19C2 and SCS12C3 were able to produce phosphatase. Biochemical characterization was carried out to analyse the various biochemical properties of bacterial isolates (Athira and Anith 2020).
Table 3. Biochemical characterization of bacterial isolates GFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2, SCS03C1, SCS06C1, SCS07C3, SCS12C1,SCS12C2, SCS12C3 and SCS12C5 using Vitek 2. (Sr. 1 to 23).
| Sr | Biochemical tests | GFS | GFS | GFS | GFS | GFS | GFS | GFS | SCS | SCS | SCS | SCS | SCS | SCS | SCS |
| 01 | 03 | 01 | 15 | 16 | 16 | 19 | 03 | 06 | 07 | 12 | 12 | 12 | 12 | ||
| C1 | C1 | C1 | C2 | C1 | C2 | C2 | C1 | C1 | C3 | C1 | C2 | C3 | C5 | ||
| 1 | Ala-Phe-Pro-Arylamidase | – | + | + | – | – | + | – | – | – | – | – | – | + | – |
| 2 | Hydrogen Sulfide | (+) | + | + | – | – | – | – | – | – | – | – | + | – | – |
| 3 | Beta-Glucosidase | – | – | – | + | – | + | + | + | – | – | – | – | + | + |
| 4 | L-Proline Arylamidase | – | – | – | – | – | + | – | – | – | + | – | – | + | – |
| 5 | Saccharose/Sucrose | + | – | + | – | – | – | + | + | – | – | – | – | – | + |
| 6 | L-Lactate Alkalinisation | + | – | – | + | + | – | + | – | + | + | + | – | – | + |
| 7 | Glycine Arylamidase | – | – | – | – | – | + | – | – | – | + | – | – | + | – |
| 8 | O/129 Resistance (Comp.Vibrio.) | + | + | + | + | + | – | + | + | + | + | + | + | – | – |
| 9 | Adonitol | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 10 | Beta-N-Acetyl-Glucosaminidase | + | + | + | + | – | + | – | – | – | – | – | – | + | – |
| 11 | D-Maltose | + | + | + | + | – | + | + | + | + | – | – | + | + | + |
| 12 | Lipase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 13 | D-Tagatose | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 14 | Alpha-Glucosidase | + | + | + | + | – | + | + | + | + | – | – | + | + | + |
| 15 | Ornithine Decarboxylase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 16 | Glu-Gly-Arg-Arylamidase | – | – | – | – | – | + | – | – | – | – | – | – | + | – |
| 17 | L-Pyrrolydonyl-Arylamidase | + | + | + | + | – | + | + | + | + | + | – | + | + | + |
| 18 | Glutamyl Arylamidase pNA | – | – | – | – | – | + | – | – | – | – | – | – | + | – |
| 19 | D-Mannitol | – | – | – | – | – | – | + | + | + | – | – | – | – | + |
| 20 | Palatinose | – | – | – | – | – | – | + | + | + | – | – | – | – | + |
| 21 | D-Trehalose | + | + | + | + | – | – | + | + | + | – | – | + | – | + |
| 22 | Succinate Alkalinization | – | – | – | – | + | – | – | – | – | – | + | – | – | – |
| 23 | Lysine Decarboxylase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
Table 4. Biochemical characterization of bacterial isolates GFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2, SCS03C1, SCS06C1, SCS07C3, SCS12C1, SCS12C2, SCS12C3 and SCS12C5 using Vitek 2. (Sr. 24 to 47)
| Sr. | Biochemical tests | GFS | GFS | GFS | GFS | GFS | GFS | GFS | SCS | SCS | SCS | SCS | SCS | SCS | SCS |
| 1 | 3 | 11 | 15 | 16 | 16 | 19 | 3 | 6 | 7 | 12 | 12 | 12 | 12 | ||
| C1 | C1 | C1 | C2 | C1 | C2 | C2 | C1 | C1 | C3 | C1 | C2 | C3 | C5 | ||
| 24 | L-Malate Assimilation | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 25 | L-Arabitol | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 26 | D-Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 27 | D-Mannose | + | + | – | – | + | + | + | + | + | + | + | – | + | – |
| 28 | Tyrosine Arylamidase | + | – | – | – | + | + | + | + | + | + | + | – | + | + |
| 29 | Citrate (Sodium) | – | – | – | – | + | – | + | + | + | + | + | – | – | + |
| 30 | Beta-N-Acetyl-Galactosaminidase | – | – | – | – | – | + | – | – | – | – | – | – | + | – |
| 31 | L-Histidine Assimilation | – | – | – | – | – | – | – | – | – | – | + | – | – | – |
| 32 | Ellman | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 33 | D-Cellobiose | – | – | – | – | + | + | + | + | + | – | + | – | + | + |
| 34 | Gamma-Glutamyl-Transferase | – | – | – | – | – | + | + | + | + | – | – | – | + | – |
| 35 | Beta-Xylosidase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 36 | Urease | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 37 | Malonate | – | – | – | – | + | – | – | – | – | – | – | – | – | – |
| 38 | Alpha-Galactosidase | – | – | – | – | – | + | (-) | – | + | – | – | – | + | + |
| 39 | Courmarate | + | + | + | + | + | – | – | – | + | + | + | + | – | – |
| 40 | L-LACTATE Assimilation | – | – | – | – | – | – | – | – | + | – | + | – | – | + |
| 41 | Beta-Galactosidase | – | – | – | – | – | (-) | – | – | + | – | – | – | (-) | + |
| 42 | Fermentation/ Glucose | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 43 | BETA-Alanine Arylamidase Pna | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 44 | D-Sorbitol | – | – | – | – | – | – | – | – | – | – | – | – | – | + |
| 45 | 5-Keto-D-Gluconate | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 46 | Phosphatase | + | + | + | + | – | + | + | – | – | – | – | – | + | – |
| 47 | Beta-Glucuronidase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
Biochemical characterization using Vitex 2 compact was done to analyse biochemical characterization of bacterial isolates. Isolates GFS01C1, GFS03C1, GFS11C1, GFS15C2, GFS16C1, GFS16C2, GFS19C2, SCS03C1, SCS07C3, SCS12C1, SCS12C2, SCS12C3, SCS12C5 and SCS06C1 were not able to utilise adonitol, D-tagatose and L-arabitol. None of the bacterial isolates were able to produce lipase, ornithine decarboxylase, lysine decarboxylase, Beta-xylosidase, urease, beta-glucuronidase 5-Keto-D-Gluconate and beta-alanine arylamidase. None of the bacterial isolates were able to assimilate of L- malate. All the bacterial isolates were negative for Ellman reaction and fermentation of glucose. All the bacterial isolate were able to acidify D-glucose which basically leads to drop in pH of the medium. Similar pattern was observed during quantitative analysis of phosphate solubilization
Cluster analysis was performed to study the biochemical relatedness based of biochemical tests among the bacterial isolates, using PAST. ver. 4.07b.(Hammer et al. 2001; Athira and Anith 2020).
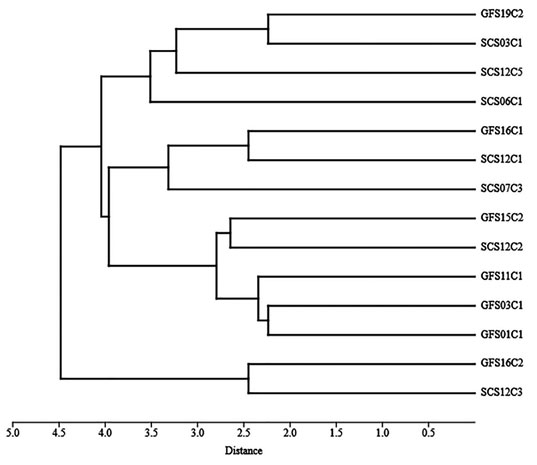
Using cluster analysis (figure 4) by euclidean distance method based on biochemical characterization isolates GFS16C2 & SCS12C3 have distinct characters than other isolates. GFS01C1&GFS03C1 and GFS19C2&SCS03C1 are more related as compared with other isolates.Individual bacteria cells specialize into unique tasks randomly and independently of one another in some bacteria species by random variations in each cell’s metabolic processes.In Bacillus subtilis, for example, whether a cell makes and secretes protease is decided at random(Bettenworth et al. 2019; Li et al. 2021). However, the use of 16S rDNA amplicon sequencing can addressthis drawback(Liu et al. 2021).
Figure 4: Clustering of the bacterial isolates using based on Euclidean distance tree constructed using PAST.
CONCLUSION
The findings of the present study indicate the quantitative analysis of phosphate solubilizing capability and IAA synthesis by bacterial isolates. The current study attempted to identify plant growth enhancing microorganisms that may be harnessed to increase plant development. Several phosphate solubilizers and IAA makers also produced siderophores and HCN, suggesting that these organisms are capable of biocontrol. As a result, it can be deduced that some of the screened isolates have a strong capacity to operate as PGPR. Identifying microorganisms and using seed tests can also help us understand dynamics.
Conflict of Interests: Authors declare noconflicts of interests to disclose.
Data Availability Statement: The database generated and /or analysed during the current study are not publicly available due to privacy, but are available from the corresponding author on reasonable request.
REFERENCES
Arora, N. K. and Verma, M. (2017). Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech, 7, 1-9.
Backer, R., Rokem, J. S., Ilangumaran, G., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Frontiers in plant science, 9, 1473.
Bandopadhyay, A., Bhattacharya, S. K. and Das, N. (2019). Biocontrol and growth promoting potential of eight PGPFs on jute and sunnhemp. Journal of Soils Crops, 29, 243-250.
Basu, A., Prasad, P., Das, S. N., et al. (2021). Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: recent developments, constraints, and prospects. Sustainability, 13, 1140.
Bettenworth, V., Steinfeld, B., Duin, H., et al. (2019). Phenotypic Heterogeneity in Bacterial Quorum Sensing Systems. Journal of Molecular Biology, 431, 4530-4546.
Bhat, M. A., Kumar, V., Bhat, M. A., et al. (2020). Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Frontiers in Microbiology, 11, 1952.
Dipak, S. and Abhijit, H. (2005). Physical and chemical methods in soil analysis, New Age International (P) Limited, Publishers.
Fahmy, T. (2016). XLSTAT. 2016.02.28451 ed.: Addinsoft.
Hammer, Ø., Harper, D. A. and Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. 4.07b ed.: Palaeontologia electronica.
Cappuccino, J. and Welsh, C. (2017). Microbiology: A Laboratory Manual, Pearson Education.
Joshi, G., Kumar, V. and Brahmachari, S. K. (2021). Screening and identification of novel halotolerant bacterial strains and assessment for insoluble phosphate solubilization and IAA production. Bulletin of the National Research Centre, 45, 1-12.
Kaneriya, J. P., Pattani, V. B. and Joshi, K. (2021). Screening and Isolation of Plant Growth-Promoting Bacteria from Forest and Coastal Regions of Saurashtra, Gujarat.Bioscience Biotechnology Research Communications, 14, 410-415.
Karthika, S., Varghese, S. and Jisha, M. S. (2020). Exploring the efficacy of antagonistic rhizobacteria as native biocontrol agents against tomato plant diseases. 3 Biotech, 10, 320.
Kenneth, O. C., Nwadibe, E. C., Kalu, A. U., et al. (2019). Plant growth promoting rhizobacteria (PGPR): a novel agent for sustainable food production. American Journal of Agricultural and Biological Sciences, 14, 35-54.
Khatoon, Z., Huang, S., Rafique, M., et al. (2020). Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. Journal of Environmental Management, 273, 111118.
Kour, D., Rana, K. L., Yadav, N., et al. (2019). Rhizospheric microbiomes: biodiversity, mechanisms of plant growth promotion, and biotechnological applications for sustainable agriculture. Plant Growth Promoting Rhizobacteria for Agricultural Sustainability. Springer,19-65.
Kumar, A., Patel, J. S., Meena, V. S., et al. (2019). Recent advances of PGPR based approaches for stress tolerance in plants for sustainable agriculture. Biocatalysis and agricultural biotechnology, 20, 101271.
Kumar, S., Sindhu, S. S. and Kumar, R. (2021). Biofertilizers: An ecofriendly technology for nutrient recycling and environmental sustainability. Current Research in Microbial Sciences, 100094.
Lebrazi, S., Fadil, M., Chraibi, M., et al. (2020a). Screening and optimization of indole-3-acetic acid production by Rhizobium sp. strain using response surface methodology. 18, 1-10.
Lebrazi, S., Niehaus, K., Bednarz, H., et al. (2020b). Screening and optimization of indole-3-acetic acid production and phosphate solubilization by rhizobacterial strains isolated from Acacia cyanophylla root nodules and their effects on its plant growth. Journal of Genetic Engineering and Biotechnology, 18, 71.
Li, E., Ryo, M., Kowalchuk, G. A., et al. (2021). Rapid evolution of trait correlation networks during bacterial adaptation to the rhizosphere. Evolution, 75, 1218-1229.
Liu, M., West, S. A. and Cooper, G. A. (2021). Relatedness and the evolution of mechanisms to divide labor in microorganisms. Ecology and Evolution, 1–15.
Lorck, H. (1948). Production of Hydrocyanic Acid by Bacteria. Physiologia Plantarum, 1, 142-146.
Marra, L. M., de Oliveira-Longatti, S. M., Soares, C. R. F. S., et al. (2019). The Amount of Phosphate Solubilization Depends on the Strain, C-Source, Organic Acids and Type of Phosphate. Geomicrobiology Journal, 36, 232-242.
Mony, C., Vandenkoornhuyse, P., Bohannan, B. J., et al. (2020). A landscape of opportunities for microbial ecology research. Frontiers in Microbiology, 11, 2964.
Morales-Cedeno, L. R., del Carmen Orozco-Mosqueda, M., Loeza-Lara, P. D., et al. (2021). Plant growth-promoting bacterial endophytes as biocontrol agents of pre-and post-harvest diseases: Fundamentals, methods of application and future perspectives. Microbiological Research, 242, 126612.
Pereira, L. B., Andrade, G. S., Meneghin, S. P., et al. (2019). Prospecting Plant Growth-Promoting Bacteria Isolated from the Rhizosphere of Sugarcane Under Drought Stress. Current Microbiology, 76, 1345-1354.
Prasad, M., Srinivasan, R., Chaudhary, M., et al. (2019). Plant growth promoting rhizobacteria (PGPR) for sustainable agriculture: perspectives and challenges. PGPR amelioration in sustainable agriculture. Elsevier,129-157.
Rawat, P., Shankhdhar, D. and Shankhdhar, S. (2020). Plant growth-promoting rhizobacteria: a booster for ameliorating soil health and agriculture production. Soil Health. Springer,47-68.
Schwyn, B. and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry, 160, 47-56.
Sharma, P., Sharma, M. M. M., Kapoor, D., et al. (2020). Role of microbes for attaining enhanced food crop production, Springer Nature Singapore Pte Ltd.
Shastri, B., Kumar, R. and Lal, R. J. (2020). Isolation, Characterization and Identification of Indigenous Endophytic Bacteria Exhibiting PGP and Antifungal Traits from the Internal Tissue of Sugarcane Crop. Sugar Tech, 22, 563-573.
Slama, H. B., Cherif-Silini, H., Chenari Bouket, A., et al. (2019). Screening for Fusarium Antagonistic Bacteria From Contrasting Niches Designated the Endophyte Bacillus halotolerans as Plant Warden Against Fusarium. Frontiers in Microbiology, 9, 3236.
Suresh, A., Sameera, P., Chapla, J., et al. (2020). Plant Growth-Promoting Rhizobacteria (PGPR) Activity in Soil. Microbes in Agriculture and Environmental Development. CRC Press,165-182.
Swarnalakshmi, K., Yadav, V., Tyagi, D., et al. (2020). Significance of plant growth promoting rhizobacteria in grain legumes: Growth promotion and crop production. Plants, 9, 1596.
Tabassum, B., Khan, A., Tariq, M., et al. (2017). Bottlenecks in commercialisation and future prospects of PGPR. Applied Soil Ecology, 121, 102-117.
Vassilev, N., Vassileva, M., Martos, V., et al. (2020). Formulation of microbial inoculants by encapsulation in natural polysaccharides: focus on beneficial properties of carrier additives and derivatives. Frontiers in plant science, 11, 270.


