1Natural Products Chemistry and Pharmacology Programme, Institute of Bioresources and Sustainable Development, Imphal-795001, Manipur, India
2Distributed Information Sub-Centre, Institute of Bioresources & Sustainable Development, Imphal -795001, Manipur, India
3Natural Product Chemistry, Institute of Advanced Study in Science & Technology, Guwahati -781035, Assam, India.
4Department of Pharmaceutical Sciences, Dibrugarh University, Dibrugarh, Assam 786004, India
5Microbial Resources Programme, Institute of Bioresources and Sustainable Development, Imphal-795001, Manipur, India
6Department of Pharmacology, Jawaharlal Nehru Institute of Medical Sciences, Imphal 795001, Manipur, India
7Plant Systematic Conservation Laboratory, Institute of Bioresources and Sustainable Development, Imphal-795001, Manipur, India
Corresponding author Email: b_thongam07@yahoo.com
Article Publishing History
Received: 30/09/2018
Accepted After Revision: 24/10/2018
Diabetes mellitus is rapidly emerging creating major health problem worldwide. Though synthetic drugs are available, due to their association with side effect, there is always interest for search of herbal formulation. Quercus serrata is a plant used by traditional healers of Manipur, India as a folk remedy to treat diabetes mellitus. The objective of this study is bioassay guided isolation of á-glucosidase inhibitory compound from the leaves of Quercus serrata and to check the postprandial antihyperglycaemic effect of the isolated compound in STZ-induced diabetic albino mice. And to perform molecular docking studies to predict the binding interaction of the isolated compounds with á-glucosidase.The methanolic extract of Quercus serrata leaves was screened for á-glucosidase inhibitory activity and fractionated into n-butanol, ethyl acetate and water fraction to isolate the active compound. Quantification of isolated compound was done by HPLC-PDA. Postprandial antihyperglycaemia effect was check in normal and STZ (100mg/kg) + nicotinamide (120mg/kg) induced diabetic mice after sucrose load (2g/kg). Molecular docking study was done using AutoDock 4.2.6. Rosmarinic acid was identified as the active compound present in Quercus serrata leaves responsible for the inhibition of á-glucosidase with IC50 0.23 ± 0.02 μgmL-1 (0.636µmolmL-1). Postprandial blood sugar and Area Under Curve were significantly (p<0.05) reduced by treatment with rosmarinic acid in normal and diabetic mice. Additionally, in silico docking study elaborated the possible binding between rosmarinic acid and á-glucosidase. Above finding confirmed the anti-diabetic potential of traditionally used Quercus serrata leaves and the isolated compound, rosmarinic acid.
Á-Glucosidase, Diabetes Mellitus, Molecular Docking, Postprandial Hyperglycemia, Quercus Serrata, Rosmarinic Acid
Chanu M. B, Labala R. K, Sheikh Y, Borah J. C, Ghosh S. K, Sahoo D, Singh O. J, Shakya A, Thongam B. Bioassay Guided Isolation of á-Glucosidase Inhibitory Compound, in Vivo Postprandial Anti Hyperglycemia and Docking Study of the Isolated Compound from the Leaves of The Methanolic Extract of Quercus Serrata. Biosc.Biotech.Res.Comm. 2018;11(4).
Chanu M. B, Labala R. K, Sheikh Y, Borah J. C, Ghosh S. K, Sahoo D, Singh O. J, Shakya A, Thongam B. Bioassay Guided Isolation of á-Glucosidase Inhibitory Compound, in Vivo Postprandial Anti Hyperglycemia and Docking Study of the Isolated Compound from the Leaves of The Methanolic Extract of Quercus Serrata. Biosc.Biotech.Res.Comm. 2018;11(4). Available from: https://bit.ly/2kL9OCr
Introduction
Diabetes mellitus and the spectrum of complications associated with it impose uncertain healthcare challenges and economic burdens to the global population. The catastrophic prevalence of diabetes mellitus, predominantly type 2 diabetes, has become a global healthcare problem affecting 425 million people worldwide, (Zimmet et al., 2014, IDF Diabetes Atlas, 2017). In India, it is reaching extreme epidemic level. A recent study revealed that prolongation of diabetes leads to neuropathy (24.6%), the most common complication, followed by cardiovascular complications (23.6%), renal problems (21.1%), and retinopathy (16.6%) (Kaveeshwar and Cornwall, 2014).
Diabetes is a life style disease and food habit plays a major role in the management of diabetes. Control of postprandial hyperglycemia is also one of the measures to control severity of the disease. Acarbose, miglitol and voglibose are available clinically used drug to control postprandial absorption of glucose by inhibiting the enzyme á-glucosidase and á-amylase. Although substantial quantum of efforts has been made towards conquering the disease, it is still imperative to put rigorous effort in research to mitigate the disease.There is always search for new herbal formulation to avoid the side effect associated with the synthetic drug, (Polonsky, 2012, Nakatsu et al., 2017, Brito-Arias et al.,2018).
Quercus serrata Murray is widely used by traditional healers of Manipur, India as a folk remedy to treat diabetes mellitus (Sheikh et al 2015). No elaborate study has been reported on the medicinal use of Quercus serrata leaves and the chemical constituent present. So, the present study is aim at bioassay guided isolation of the active constituent responsible for the inhibition of the enzyme á-glucosidase, checking its antihyperglycaemic activity and to do molecular docking study of the isolated compound to predict the possible binding interaction of the isolated compound with the enzyme, á-glucosidase.
Materials and Methods
Leaves of Quercus serrata, Murray were collected from Kangla Siphai, Manipur, India and authenticated by Dr. Biseshwori Thongam, Scientist D, Pant Taxonomist, IBSD, Manipur. Voucher specimen (No. IBSD/M-202) was deposited in the IBSD herbarium. Analytical or HPLC grade organic solvents (Merck Millipore, India) were used for experiments. For column chromatography, 100-200 mesh size silica gel (Merck) was used. á-Glucosidase (Maltase, EC 3.2.1.20), p-nitrophenyl-á-D-glucopyranoside and streptozotocin were purchased from Sisco Research Laboratory. HPLC (Shimadzu LC-20AD) was performed using Photo diode array (PDA) detector. NMR spectra were recorded on a Bruker Avance 500 MHz instrument with TMS as an internal standard. Chemical shifts were expressed in á values. Agilent 6520 Accurate mass Q-TOF/LC-MS was used to determine molecular weight. Absorbance was measured by Thermo Scientific Multiskan spectrometer.
Extraction and isolation of active compound
Air-dried leaves of Quercus serrata (3.5 kg) were extracted three times with methanol (MeOH) (15 L). Then 150 mL water was added to the concentrated methanolic extract (3.0 L) to make hydro-alcoholic solution. This solution was washed with 3.0 L of petroleum ether to remove fatty matter present in the leaves and then concentrated to yield 400 g of dried extract. A suspension was prepared from this using 500 mL of water and fractionated into ethylacetate , n-butanol and water to yield 152 g of ethyl acetate fraction, 103 g of butanol fractions and 135g of water fraction respectively. The butanol fraction was found to be most active and was subjected to silica gel column chromatography with increasing polarity: petroleum ether-chloroform 1:1 (Fr 1, 1 L), chloroform (Fr 2, 1 L), and chloroform – methanol 9:1 (Fr 3, 2 L), chloroform – methanol 4:1 (Fr 4, 2 L), chloroform – methanol 1:1 (Fr 5, 2.5 L) and chloroform – methanol 3:7 (Fr 6, 2.5 L). The sub-fraction 6 (SFr 6) was found to be most active in inhibition of á-glucosidase. Further, SFr 6 was subjected to semi- preparative HPLC to yield compound 1 (rosmarinic acid).
Analytical and semi-preparative HPLC
Shimadzu LC-20AD with PDA detector (SPD-M20A) fitted with either an analytical (CAPCELL PAK C18 MGII S5, 5µ C18 250 × 4.6 mm) or semi-preparative (CAPCELL PAK C18 MGII S5, 5µ C18 250 × 10.00 mm) column was used for HPLC analysis. A solution of 10 mg/mL of enriched fraction (SFr 6) was prepared in HPLC grade water and methanol (7:3) and filtered through 0.45µm Millex-HN syringe filter. A 20µL or 200µL aliquot of filtered sample solution was injected for analysis for analytical and semi preparative separation respectively. The mobile phase used consisted of 1% acetic acid in water (solvent A) and methanol (solvent B) following linear gradient over a total run time of 23min: initially a linear increase of B up to 100% in 21 min and back to 100% A in 23 min. Flow rate was 1 mL/min and 3 mL/min for analytical and semi-preparative respectively. Individual peaks eluting from the column were carefully collected and the solvents were evaporated in a rotary vacuum evaporator. Instrument control and data handling were performed with the LC solution software on a PC.
Quantitative analysis by HPLC-PDA
Quantitative analysis was done by analytical HPLC in the protocol described above. Standard stock solution of 1mg/mL of rosmarinic acid was prepared and then diluted to yield the desired test concentrations. Quantitative estimation of the major active compound present in the methanolic extract, n-butanol fraction and enriched fraction (SFr6) was done using calibration curve of the standard solution and plotted concentration versus area using excel 2007 (Sheikh et al., 2016).
á-Glucosidase inhibitory assay
A 0.5U/mL stock solution of readily available á-glucosidase (from Saccharomyces cerevisiae) was prepared in 0.1M phosphate buffer (pH 6.8) and diluted with the same buffer to desired test concentration. Stock solution of plant extract, fractions and compounds were prepared in dimethyl sulfoxide (DMSO) and diluted for assay to the required concentration with same buffer maintaining DMSO concentration below 1% v/v. á-glucosidase inhibitory activities were determined spectrophotometrically based on an earlier reported method by using p-nitrophenyl-á-D-glucopyranoside as substrate (Laishram et al., 2014) Acarbose was used as positive control and the uninhibited enzyme was taken as negative control (DMSO control). The assay was performed in three independent experiments.
Experimental animals
Swiss albino mice (25-30 g) were used in all the experiment and were procured from Regional Institute of Medical Sciences (RIMS), Manipur, India. Ethical clearance was obtained from the Institutional Animals Ethical Committee (Approval No.–RIMS.171/IAEC/2011) prior to the experiments. Animals were acclimatized in laboratory condition for 7 days in polypropylene cages lined with husk under standard environmental conditions. Animals had free access to water and were fed on pelleted diet.
Acute oral toxicity study
The acute toxicity study was performed as per Organisation for Economic Co-operation and Development (OECD) guideline no. 423 (Annexure-2c) adopted by the Committee for the purpose of control and supervision of experiments on animals (CPCSEA), Government of India (Veeraraghavan, 2000). Mice were kept under observation for 14 days with special attention for the first 24 h after dosing.
Induction of diabetes
Diabetes was induced in mice by single intraperitoneal (i.p.) injection of nicotinamide (120mg/kg i.p.) and then 100 mg/kg b.w. of streptozotocin (freshly dissolved in 0.1M citrate buffer, pH 4). Mice showing marked hyperglycemia (fasting blood glucose ≥ 250 mg/dl) at 48 h after streptozotocin treatment were considered as diabetic and selected for the study, (Nakatsu et al., 2017)
Effect of rosmarinic acid on postprandial hyperglycemia
Both normal and diabetic mice were fasted overnight and divided into five groups (n=6). Group I served as normal control and received vehicle, group II served as positive control and received acarbose (10 mg/kg b.w.). Group III, IV and V received rosmarinic acid (5, 10 and 20 mg/kg) suspended in gum acacia (2% w/v) (Chandramohon et al., 2015). All groups received 2 g/kg b.w. sucrose orally along with their respective test samples (Miura et al., 2004). Blood glucose levels were estimated from the tail vein from each group before (0 min) and at 30, 60 and 120 min after sucrose load. Blood glucose was measured by using Accu-Check Active, Roche Diagnostic Mannheim, Germany. AUC was calculated by using the software, GraphPad Prism 5.
In silico structure prediction of á-glucosidase
S. cerevisiae á-glucosidase protein sequence (MAL12, Accession Number: P53341) was retrieved from UniProt (www.uniprot.org). NCBI CDD (Marchler-Bauer et al., 2015) was used to find out the conserved domain and other important catalytic sites. Structural 3D model for the protein was built using automated comparative protein modeling server SWISS-MODEL (https://swissmodel.expasy.org). Homology based search was performed by the server in Protein Data Bank (PDB) and SWISS-MODEL template library (SMTL) repositories for template structures (Biasini et al., 2014). Suitable template with similar biological property and highest similarity was selected for creating the 3D model. Further checking accuracy of the newly generated structure was done by using PROCHECK (Laskowski et al.,1993) and ProSA-web (Sippl, 1993; Wiederstein and Sippl, 2007) for evaluation of Ramachadran plot.
Docking study
3D structure of rosmarinic acid (PubChem CID: 5281792) was obtained from NCBI PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Surface topology was calculated using CASTp server (Dundas et al., 2016) for potential binding pockets in the structural model of á-glucosidase. Initially whole protein was considered for docking. Polar hydrogen atoms and Gasteiger charge were added to the protein molecule. Grid size of 96 Å × 120 Å × 110 Å with a spacing of 0.619 Å was set to cover the entire protein. Lamarckian genetic algorithm search was considered to generate 100 conformations and rest of the parameter was set to default. Later, a cluster analysis was carried out by considering the root mean square deviation of conformations to identify major occupied binding sites in the protein. On the basis of major CASTp pockets and top AutoDock clusters information, a smaller grid box, 62 Å × 60 Å × 70 Å with a spacing of 0.464 Å was determined for the second set of docking simulation. All other parameters and procedureof the docking program were kept as earlier.
The best docked conformation of rosmarinic acid was selected based on lowest binding energy. Minimization of the docked complex was then performed using CHARMm force-field with smart minimizer (2000 steps) by considering Generalized Born with Molecular Volume (GBMV) implicit solvent model. Binding free energy (ΔG) of the docked complex was calculated by using the same GBMV model. Implicit Distance-Dependent Dielectric model was used to calculate total and individual interaction energy (IE) possessed by the amino acid residues. Molecular graphics, analysis and depiction were performed using UCSF Chimera (Pettersen et al., 2004) and BIOVIA Discovery Studio Visualizer Version 4.5 (http://www.3dsbiovia.com).
Statistical analysis
Results were expressed as Mean ± S.D., where n=6. Differences among data were determined using one way ANOVA followed by Tukey’s Multiple Comparison test (Graph Pad Prism software, version 7). *p<0.05 was considered statistically significant.
Results And Discussion
Quantitative HPLC analysis: Rosmarinic acid (Fig. 1) was found to be the major compound present in the enriched fraction Sfr 6. Fig. 2 showed the HLPC chromatogram of rosmarinic acid in methanolic extract, n-butanol fraction, enriched fraction (SFr 6) as well as that of isolated rosmarinic acid. The calibration curve (Fig. 3) of the standard rosmarinic acid was found to be linear (r2=0.993) in the concentration range of 0.01 to 0.09 mg/ml. 54.28±4.14 mg, 239±1.21 mg and 426±8.36 mg of rosmarinic acid was present per 1gm each of methanol extract, n-butanol fraction and enriched fraction (SFr 6) respectively.
 |
Figure 1: Structure of Rosmarinic acid |
 |
Figure 3: Calibration curve of standard rosmarinic acid as obtained from HPLC chromatogram. The graph is plotted, Area against Concentration (mg/ml) from the range 0.01 to 0.09mg/ml. |
Rosmarinic acid NMR interpretation
Compound 1 (Fig. 1) gave a molecular ion peak in HRMS at m/z = 360.3212 [M+H]+ corresponding to the molecular formula C18H16O8. The 13C NMR in CD3OD indicated the presence of signals attributed to all 18 carbon atoms. One ester carbonyl at ä = 167.06 displayed HMBC correlation with H-7’ (ä 7.58), coupled to a doublet of J = 14.5 Hz at ä 6.34 indicative of a cinnamoyl ester. Another carbonyl at ä 172.06 showed HMBC correlation with signal of H-7 (ä 3.13, H-7a and ä 3.05, H-7b), coupled to a methene signal at ä 5.20, agreeing with the presence of a phenyl lactic acid unit. Two ABX systems were noted in the aromatic region of the 1H NMR vouching for 3,4-dihydroxylation pattern of both the rings. The linkage of the cinnamoyl ester to the phenyl lactic acid moiety was inferred from HMBC correlations displayed by the H-8 signal to both the carbonyl peaks. Signals at ä 6.98 (1H, d, J = 2.0 Hz, H-6’) and ä 7.06 (1H, dd, J = 8.5, 2.0 Hz, H-2’) showing meta coupling between them and ortho coupling between H-2’ and H-3’ with signal at ä 6.79 (1H, d, J = 8.5 Hz). Another ABX system with signal at ä 6.64 (1H, d, J = 2.0 Hz, H-2), ä 6.70 (1H, dd, J = 9.0, 2.0 Hz, H-6) and ä 6.69 (1H, d, J = 9.0 Hz, H-5) is observed. All signals of 1H and 13C, and the observed HMQC, 1H-1H COSY and HMBC correlations are in good agreement with literature reports on rosmarinic acid.
1H NMR (500 MHz, CD3OD, ä ppm): 7.58 (1H, d, J = 14.5 Hz, H-7/), 7.06 (1H, dd, J = 2.0, 8.5 Hz, H-2/), 6.98 (1H, d, J = 2.0 Hz, H-6/), 6.79 (1H, d, J = 8.5 Hz, H-3/), 6.70 (1H, dd, J = 2.0, 9.0 Hz, H-6), 6.69 (1H, d, J = 9.0 Hz, H-5), 6.64 (1H, d, J = 2.0 Hz, H-2), 6.34(1H, d, J = 14.5 Hz, H-8/), 5.20 (1H, m, H-8), 3.13 (1H, m, H-7a) and 3.05 (1H, m, H-7b).13C NMR (125 MHz, CD3OD, ä ppm): 172.06 (C-9), 167.06 (C-9/), 148.34 (C-4/), 146.35 (C-7/), 145.41 (C-5/), 144.76 (C-4/), 143.89 (C-3), 127.84 (C-1), 126.25 (C-1/), 121.77 (C-2), 120.40 (C-6/), 116.18 (C-6), 115.10 (C-5), 114.89 (C-3/), 113.81 (C-8/), 112.99 (C-2/), 73.18 (C-8) and 36.51 (C-7). (Supp Fig. 1a, 1b)
Oral acute toxicity study
No fatality and ethological changes were observed when administered a dose of 2000 mg/kg b.w. of the methanolic extract. No changes in behavioral pattern were observed in the mice in oral toxicity study. So, Quercus serrata leaves were non toxic upto a dose of 2000mg/kg.
In vitro ä-glucosidase inhibitory effect
Blood glucose levels are highly affected by the saccharides contained in food which are converted into glucose by the actions of digestive enzymes like ä-glucosidase. Carbohydrates like sucrose are hydrolysed to monosaccharides (glucose and fructose) by ä-glucosidase thereafter caused an increased in blood glucose. The methanolic extract of Quercus serrata leaves, ethyl acetate fraction, n-butanol fraction, enriched fraction (SFr 6) and rosmarinic acid potentially inhibited ä-glucosidase, and the result is shown as table 1. The above result signifies Quercus serrata as a promising plant for postprandial management of diabetes mellitus type 2. Moreover, rosmarinic acid showed greater potential in inhibition of ä-glucosidase in vitro when compared to that of acarbose.
| Table 1: Half maximum inhibitory concentration (IC50) of the tested samples on á-glucosidase | |
| Samples | IC50 (μgmL-1) |
| MeOH extract | 1.84 ± 0.11 |
| Ethylacetate fraction | 2.83 ± 0.07 |
| n-butanol fraction | 1.15 ± 0.06 |
| Water fraction | No inhibition up to 500 μgmL-1 |
| n-butanol Sub fraction (SFr 6) | 0.66 ± 0.01 |
| Rosmarinic acid | 0.23 ± 0.02(0.636 µmolmL-1) |
| Acarbose | 78.2 ± 0.17 |
| Each value is the mean ± SD (n=3). | |
Postprandial antihyperglycaemic effect of rosmarinic acid
Rosmarinic acid is a polyphenol compound. It is an ester of caffeic acid and 3,4-dihydroxyphenyllactic acid (Petersen and Simmonds, 2003). The effect of rosmarinic acid on postprandial blood glucose level after sucrose load in overnight fasted normal and diabetic mice were presented in the Fig 4A & B. In diabetic mice postprandial blood glucose increased from 19.14±1.01 at 30 min to 20.81±0.62 mmol/L at 60 min and decreased to 18.50±0.51 mmol/L at 120 min. Rosmarinic acid at a dose of 20 mg/kg b.w significantly reduced the postprandial blood glucose to 15.43±0.64, 15.01±0.48 and 14.19±0.74 mmol/L at 30 min, 60 min and 120 min which value was comparable to that of acarbose 15.45±0.23, 14.42±4.01 and 14.21±1.12 mmol/L at 30 min, 60 min and 120 min respectively. Rosmarinic acid at all the doses caused significant fall in AUC (Table 2) when compared to diabetic control (56.31±1.50 mmol.min/L) indicating its effectiveness in lowering postprandial glucose absorption. There are studies which reported that medication which can flattens the peak blood glucose level postprandial can reduce the AUC (Inoue et al.1997). Postprandial blood glucose and AUC in normal rats was in consistent with that of diabetic groups.
| Table 2: AUC of postprandial glucose after sucrose load in normal and diabetes mice | ||
|
Group (mg/kg.bw) |
AUC mmol.min/L | |
| Normal rats | Diabetic rats | |
| Control | 32.15±0.23 | 56.31±1.50 |
| Acarbose(10) | 19.95±0.41** | 43.90±0.94* |
| Rosmarinic acid (5) | 23.35±0.43* | 48.82±0.57* |
| Rosmarinic acid (10) | 22.71±0.25* | 48.85±1.21* |
| Rosmarinic acid (25) | 20.20±0.21** | 44.80±1.33* |
| Each value is the mean ± SD (n=6). *p< 0.05 statistically significant comparing to control. | ||
Docking
To understand the binding interaction between rosmarinic acid and á-glucosidase, molecular docking was performed. Yeast and human glucosidase are similar in their substrate specificity, pH optimum, and inhibitor sensitivity. Thus, the yeast enzyme besides its affordability serves as a good experimental model to learn more about the structure, substrate specificity, and enzymatic mechanism of human glucosidase (Brito-Arias et al., 2018).
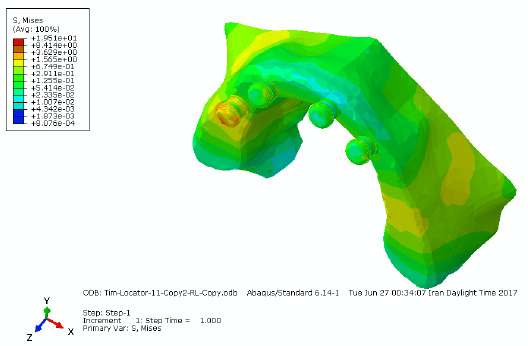
AutoDock 4.2.6 (Morris et al., 2009) incorporated in MGL Tools version 1.5.6 (The Scripps Research Institute) was used to execute the docking simulations. In this docking study á-glucosidase was treated as rigid molecule and rosmarinic acid as flexible. Due to non availability of three-dimensional crystal structure of S. cerevisiae á-glucosidase, homology study is done. Based on homology study , the protein structure of S. cerevisiae isomaltase (PDB ID: 3AJ7, Chain A at 1.3Å resolution) which got 72% similarity and 99% query coverage when compare to S. cerevisiae á-glucosidase, MAL12 was chosen for our study as the best template to build the 3D structure for S. cerevisiae á-glucosidase. (Fig 5)
Molecular docking study found that Domain 1 of á-glucosidase resembled á-amylase catalytic domain with a gap of 42 amino acids, named here as AamyGap42 (Fig. 6a). These AamyGap42 site residues are shaped as á-helices near the entry point amino acid residues (HIS279, THR307, SER308, PRO309, PHE310, PHE311, and ARG312) of the active site pocket of the enzyme (Yamamoto et al., 2010).
According to Benkert et al 2009, GMQE score which is expressed between 0 and 1 is a score where higher value indicate increased stability of the predicted structure. GMQE score for our final 3D model was found to be 0.92 which imply good model accuracy. The evaluation of the obtained models was done using the PROCHECK program through Ramachandran plot. Ramachandran plot revealed 88.8% residues were within the most favoured regions, 10.6% of residues were in additional allowed regions, 0.4% residues were in generously allowed regions, and only 0.2% residues were found in the disallowed regions. The percentage of residues in most favorable regions showed the quality of protein models. The above result obtained indicated that the model developed was qualified to be used for molecular docking process.
Surface topology analysis, was done to know the potential binding pockets in the structural model of á-glucosidase. Surface topology analysis on the protein have identified two major pockets, named here as Pocket 1 and Pocket 2 with volume of 431.6 Å3 and 384.7 Å3 respectively (Fig. 7). Two sets of docking study were performed, first by involving the whole protein, resulting in multiple conformations of the rosmarinic Acid. Root mean square deviation cluster analysis of these docked conformations showed two most populated clusters, Cluster 1 and Cluster 2 (Fig. 7). Auxiliary observations have shown that all the pockets and clusters were surrounding the AamyGap42 site with Cluster 1 directly falling under it. The catalytic and active sites of the protein were falling under Pocket 2. CASTp pockets (Pocket 1 and Pocket 2) along with AutoDock major clusters (Cluster 1 and Cluster 2) lead the target area in the protein to perform second set of docking experiment. This targeted docking study showed that rosmarinic acid did not directly bind at the active site present in Pocket 2 of the protein, instead it attached to the AamyGap42 site of Domain 1 with the lowest binding energy of -7.72 kcal/mol (Fig. 6b).
Binding details showed that rosmarinic acid interacted with 12 amino acids in the AamyGap42 site forming total 6 hydrogen bonds, two each with amino acid residue GLU402 and TYR404, and one each with LYS417 and LYS422. This interaction substantially increased the binding affinity as these amino acid residues were also found to be the major contributor of electrostatic interaction energy (IE). Moreover, the residues TYR413 and LYS410 were involved in Pi-Pi T-shaped and Pi-alkyl interactions respectively (Fig. 8a,8b). Pi-Pi T-shaped interaction between TYR413 and first benzene ring of the rosmarinic acid, Pi-Alkyl interaction with LYS410, and van der Waals interaction with PRO400, ILE401, GLU405, ASP406, VAL409, GLU414, strengthened the stability of the enzyme and rosmarinic acid complex. Binding free energy (ΔG) between the receptor and ligand was found to be -19.14 kcal/mol. The energy value suggested the possibility of inhibition of á-glucosidase by rosmarinic acid. The interaction energies (IE) between the protein and ligand analysed for each amino acid residues and overall residues showed electrostatic IE as -44.70 kcal/mol and van der Waals IE as -31.22 kcal/mol which showed good binding energy with the enzyme.
For the first time we have reported the anti-diabetic effect of Quercus serrata leaves extract and the dominant active constituent rosmarinic acid. Further, this study reported the in silico binding analysis of rosmarinic acid with the enzyme á-glucosidase of S. cerevisiae.
Conflict of interest
The authors declare no competing interest.
Acknowledgements
Authors are thankful to the Director, IBSD, Imphal and Department of Biotechnology (DBT), Govt. of India and the Director, Institute of Advanced Study in Science and technology for providing necessary infrastructure to carry out this work. Authors are also thankful to Dr. Basudeb Achari and Dr. Bikash C Pal for their valuable suggestions.
References
Benkert, P., Künzli, M. and Schwede, T. (2009) QMEAN server for protein model quality estimation. Nucleic Acids Research, 37, W510–W514. http://doi.org/10.1093/nar/gkp322.
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., Kiefer, F., Gallo, C.T., Bertoni, M., Bordoli, L. and Schwede, T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Reserch, 42, W252-W258. http://doi.org/10.1093/nar/gku340.
Brito-Arias, M., Ramírez-Hernández, P.L., García-Barrera,E.C., Perez- Venegas,M., Montiel-Valenciana, C. and Rodríguez-Pascual, L.P. (2018) Synthesis, á-Glucosidase Enzymatic Inhibition and Docking Studies of some Fused Heterocycles. Chemical & Pharmaceutical Research, 1(1)
Chandramohan, R., Pari, L., Rathinam, A. and Sheikh, B., A. (2015) Tyrosol, a phenolic compound, ameliorates hyperglycemia by regulating key enzymes of carbohydrate metabolism in streptozotocin induced diabetic rats. Chemico-biological Interactions. 229 44-54. https://doi.org/10.1016/j.cbi.2015.01.026
Dundas, J., Ouyang, Z., Tseng, J., Binkowski, A., Turpaz, Y. and Liang, J. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Research, 34, W116-W118. http://doi.org/10.1093/nar/gkl282.
Inoue, I., Takahashi, K., Noji, S., Awata, T., Negishi, K. and Katayama, S.(1997) Acarbose controls postprandial hyper-proinsulinemia in non-insulin-dependent diabetes mellitus. Diabetes Research and Clinical Practice, 36, 143-151. https://doi.org/10.1016/S0168-8227(97)00045-4.
International Diabetes Federation, (2017) Diabetes Atlas. 8th Edition. https://www.worlddiabetesfoundation.org/sites/default/files/Atlas-8e-Global-factsheet.pdf/ (accessed on 03 October 2018).
Kaveeshwar S.A.and Cornwall J. (2014) The current state of diabetes mellitus in India. Australasian Medical Journal, 7(1), 45-48. http//dx.doi.org/10.4066/AMJ.2014.1979.
Laishram, S., Sheikh, Y., Moirangthem, D.S., Deb, L., Pal, B.C., Talukdar, N.C. and Borah, J.C. (2014) Anti-diabetic molecules from Cycas pectinata Griff. traditionally used by the Maiba-Maibi. Phytomedicine, 22 (1), 23-26. https://doi.org/10.1016/j.phymed.2014.10.007.
Laskowski, R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) PROCHECK – a program to check the stereochemical quality of protein structures. J. App. Cryst. 26, 283-291.https://doi.org/10.1107/S0021889892009944.
Marchler-Bauer, A., Derbyshire, M.K., Gonzales, N.R., Lu, S., Chitsaz, F., Geer, L.Y., Geer, R.C., He, J., Gwadz, M., Hurwitz, D.I., Lanczycki, C.J., Lu, F., Marchler, G.H., Song, J.S, Thanki, N., Wang, Z., Yamashita, R.A., Zhang, D., Zheng, C. and Bryant, S.H. (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Research, 43, 222–226. http://doi.org /10.1093/nar/gku1221.
Miura, T., Itoh, C., Iwamoto, N., Kato, M., Kawai, M., Park, S.R. and Suzuki, I. (2001) Hypoglycemic activity of the fruit of the Momordica charantia in type 2 diabetic mice. Journal of Nutritional science and Vitaminology. 47(5), 340-4.
Morris, G.M., Huey, R., Lindstrom, W., Sanner, M.F., Belew, R.K., Goodsell, D.S. and Olson, A.J (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. Journal of Computational Chemistry. 30 (16), 2785-91. http://doi.org/10.1002/jcc.21256.
Nakatsu,Y., Kokubo, H., Bumdelger, B., Yoshizumi, M., Yamamotoya, T., Matsunaga, Y., Ueda, K., Inoue,Y., Inoue, M-K., Fujishiro, M., Kushiyama A., Ono, H., Sakoda H., and Asano, T. (2017) The SGLT2 inhibitor Luseogliflozin rapidly normalizes aortic mRNA levels of inflammation-related but not lipid-metabolism-related genes and suppresses atherosclerosis in diabetic ApoE KO Mice, International Journal of molecular science, 18, 1704. doi:10.3390/ijms18081704.
Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E. (2004) UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605-12. http://doi.org/10.1002/jcc.20084.
Petersen, M. and Simmonds, M.S.J. (2003) Rosmarinic acid. Phytochemistry. 62, 121-125. https://doi.org/10.1016/s0031-9422(02)00513-7.
Polonsky, K.S., (2012) The Past 200 Years in Diabetes. The New England Journal of Medicine, 367, 1332-1340. https://doi: 10.1056/NEJMra1110560.
Sheikh, Y., Chanu M.B., Biswas, D., Laishram, S., Deb, L., Talukdar , N.C. and Borah, J.C. (2015) Anti-diabetic potential of selected ethno-medicinal plants of north east India. Journal of Ethnopharmacology, 171, 37-41. https://doi.org/10.1016/j.jep.2015.05.030.
Sheikh, Y., Maibam, B.C., Talukdar, N.C., Deka D.C. and Borah, J.C. (2016) In vitro and in vivo anti-diabetic and hepatoprotective effects of edible pods of Parkia roxburghii and quantification of the active constituent by HPLC-PDA. Journal of Ethnopharmacology, 191, 21-28. https://doi.org/10.1016/j.jep. 2016.06.015.
Sippl, M.J. (1993) Recognition of Errors in Three-Dimensional Structures of Proteins. Proteins, 17(4), 355-362. http://doi.org/10.1002/prot.340170404.
Veeraraghavan, P., Expert Consultant, CPCSEA, OECD Guideline No. 423. (2000) https://ntp.niehs.nih.gov/iccvam/suppdocs/feddocs/oecd/oecd_gl423.pdf/ (accessed on 14 March 2014).
Wiederstein, M. and Sippl, M.J. (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research, 35, W407-W410. http;//doi.org/10.1093/nar/gkm290.
Yamamoto, K., Miyake, H., Kusunoki, M. and Osaki, S. (2010) Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose. The FEBS Journal, 277(20), 4205-4214. http://doi.org/10.1111/j.1742-4658.2010.07810.x.
Zimmet, P.Z., Magliano, D.J., Herman, W.H. and Shaw J.E. (2014) Diabetes: a 21st century challenge. The Lancet Diabetes and Endocrinology, 2(1), 56-64. https://doi.org/10.1016/S2213-8587(13)70112-8.