
Microbiological
Communication
Biosci. Biotech. Res. Comm. 11(3): 426-433 (2018)
Screening of indigenous active lactic acid bacteria
isolated from freshly drawn raw milk
Selvajeyanthi S
1
and Hemashenpagam N
2
1
Research Scholar, Department of Microbiology, Hindusthan College of Arts and Science
2
Professor, Department of Microbiology, Hindusthan College of Arts and Science, Coimbatore, Tamil Nadu
ABSTRACT
Main emphasis of the present study was to isolate and identify active Lactic Acid Bacteria from various raw milk.
Totally 36 freshly drawn various raw cow, buffalo, goat and sheep milk from Tirupur and Erode region in Tamil
Nadu, were collected. From the sample 56 Lactic Acid Bacteria (LAB) isolates were taken randomly. The LAB were
phenotypically identi ed and grouped based on the morphological, physiological and biochemical study. The strain
survival were also assessed under stomach acid condition like low pH and resistance to bile salt 0.3%. Antibiotic
sensitivity tests were performed for ve antibiotics. After hemolytic activity on blood agar medium the antibacte-
rial activities of the isolates were tested against two pathogenic bacteria E.coli and Staphylococcus sp., at pH 6.5 by
overlay method. The tested isolates showed invitro inhibitory zone against pathogenic bacteria. From this study we
can conclude that raw milk is good source of active lactic acid bacteria. Out of the 56 LAB isolates 28 exhibited good
probiotic properties and potentiality was characterized in future.
KEY WORDS: RAW MILK, LACTIC ACID BACTERIA, PHENOTYPICAL IDENTIFICATION, ANTIBIOTIC SENSITIVITY, ANTIBACTERIAL ACTIVITY
426
ARTICLE INFORMATION:
*Corresponding Author: selvajeyanthi@gmail.com
Received 1
st
Aug, 2018
Accepted after revision 29
th
Sep, 2018
BBRC Print ISSN: 0974-6455
Online ISSN: 2321-4007 CODEN: USA BBRCBA
Thomson Reuters ISI ESC / Clarivate Analytics USA and
Crossref Indexed Journal
NAAS Journal Score 2018: 4.31 SJIF 2017: 4.196
© A Society of Science and Nature Publication, Bhopal India
2018. All rights reserved.
Online Contents Available at: http//www.bbrc.in/
DOI: 10.21786/bbrc/11.3/11
INTRODUCTION
Ancient Indian has practice to consume freshly drawn
raw milk without boiling. They believed that raw milk
has some good property. Milk is an excellent medium
to carry an active lactic acid bacteria and buffering
capacity of milk helps to improve the survival of pro-
biotic ora in the gastrointestinal tract, (Kailasapathy
and Phillips 2008).Lactic acid bacteria tolerate high salt
concentrations as its allows the bacteria to begin metab-
olism, which produces acid that further inhibits the
growth of undesirable microorganisms (Pooja Thakkar
et al., 2015). Probiotic LAB can be a suitable alternate
of antimicrobial agents and recently found to play a

Selvajeyanthi and Hemashenpagam
positive role in mental health. Milk is one of the natural
habitats and rice source of LAB, (Delavenne et al., 2012,
Wouters et al, 2002 and Misganaw wassie and Teketay
wassie, 2016, Mokoena et al., 2016).
The LAB in milk and milk products enhance bioavail-
ability of nutrients and act as a preservative(Misganaw
wassie and Teketay wassie, 2016). Fermented and func-
tional foods and the products are crucial to the human
health(oktay Yerlikaya, 2014).Potential probiotic isolates
of Lactobacillus rhamnosus and L. plantarum were pre-
sent in indigenous goat milk (Setyawardani et al., 2011).
The probiotic L.yoghurt supplementation to worldwide
waterborne diarrhea causing Giardia infected mice
reduced the severity of Giardia infection (Geeta shukla
et al., 2010). Lactic acid bacteria produce antimicro-
bial compounds, vitamins or useful enzymes which
could help in promotion of food industry (Ashmaig
et al., 2009). Identify potent attributes to meet out cur-
rent demands of the functional food industry (Sub-
hashini, 2014).
MATERIALS AND METHODS
36 samples of raw fresh milks were collected from lac-
tating cow, buffalo, goat and sheep in the rural area
surrounding of Tirupur & Erode District. Samples were
collected using sterile centrifuge tubes and stored in
an icebox until delivery of the laboratory for analysis.
Till the analysis samples were kept in 4°C(refrigerator).
About 1ml of milk sample was mixed with 9ml of saline
[8.5g / L] to make an initial dilution [10
-1
]. The suspen-
sion was used for making suitable serial dilutions up to
10
-8
. Enumeration of LAB was determined using MRS
(Man de Rogosa Sharpe) agar and M17 agar medium
by pour plate [1ml in 15ml medium] incubated at 37 C
for 24-48 hours. After incubation colonies were chosen
based on their morphology on MRS (pH-5.7) agar plate.
The typical LAB were randomly picked up and puri ed
for further work. Simple tests such as gram staining,
catalase test, motility and sugar fermentation test were
performed for isolates.
The isolates grown in freshly prepared liquid media
and incubated overnight. After incubation the cells were
taken and then gram staining procedure was performed.
The gram reaction of the isolates was determined by
light microscopy. Catalase enzyme produced by many
microorganisms that breaks down the H
2
O
2
into water
and oxygen that releases O
2
gas bubbles. The forma-
tion of gas bubbles indicates the presence of catalase
enzyme.
2H
2
O
2
2 H
2
O + O
2
The freshly grown liquid cultures were also used for
catalase activity by dropping 3% hydrogen peroxide
solution onto 1 ml of overnight cultures and their cata-
lase activity was observed.
(Thakkar et al., 2015). MRS broth supplemented with
different Sugars (glucose, lactose and maltose) and phe-
nol red as pH indicator was inoculated with active cul-
tures at 1%, incubated at 37°C for 24 hours. The cul-
tures were identi ed based on acid and gas production
in Durham’s tube after the incubation period. To check
the growth of isolates at various pH, MRS broth sup-
plemented with different pH 2.0, 3.0, 7.0, 8.5 was pre-
pared, 1% of fresh culture was inoculated and then incu-
bated at 37ºC for 28 hours. During incubation, extent of
growth was recorded objectively based on visible turbid-
ity marked as double positive sign (++) for maximum
growth, single positive sign (+) for normal growth and
negative sign (-) for no growth. Turbidity also measured
at 620nm.Overnight active cultures were inoculated at
1% in MRS broth tubes and incubated up to 7 days at 15,
37, 45 and 55ºC. Extent of growth was visually recorded
based on intensity of turbidity. Overnight grown active
cultures were inoculated at 1% in MRS broth tubes
adjusted to various concentrations of Na Cl viz. 3.5, 6.5
and 18% (w/v) along with their respective controls. The
cultures were incubated at 37°C. After 24 hours of incu-
bation, extent of growth was recorded objectively based
on visible turbidity marked as double positive sign (++)
for maximum growth, single positive sign (+) for normal
growth and negative sign (-) for no growth.
Blood hemolysis test was carried out as per the
method of Mabrouk et al.,( 2014). As the strains were
isolated from food material, blood haemolysis test was
performed, to eradicate any chance that our isolates
may be pathogenic. It is also one of the criteria for
assessing the safety of use of probiotics as food supple-
ments. Pathogens produce highly toxic substance which
lyse the RBC and forms a clear zone around them. The
haemolytic activities of isolated strains were determined
according to (Marakoudakis et al., 2009) as follows: all
examined strains were grown in MRS broth at 37°C for
24 hours and then streaked onto Columbia agar base
plates supplemented with 5 % (v/v) whole human blood.
The plates were incubated at 37 ºC for 48 hours. Then
the clear zones and the color of haemolysis around the
growth colonies were observed. Antibiotic susceptibility
test was done using the method of Singh et al., (2014).
Probiotic strains must be sensitive to wards the anti-
biotics. There is a light risk that antibiotic resistance
probiotic strain may transfer the antibiotic resistance
genes to the pathogens via transformation in the gut.
Due to any chance resistant pathogens get introduced
into the human via food chain and cause serious prob-
lems. Sensitivity of probiotics strains towards the antibi-
otics being tested by using Kirby - Bauer disc diffusion
technique. Tetracycline, Penicillin, Vancomycin, Strep-
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS SCREENING OF INDIGENOUS ACTIVE LACTIC ACID BACTERIA ISOLATED 427
Selvajeyanthi and Hemashenpagam
tomycin and Kanamycin was used. For this process MRS
agar inoculated with LAB and disc were placed. After
the incubation period (24 hours/ 37c) and inhibition
zones were observed to determine the antibiotic resist-
ance of isolates. Antagonistic activity was carried out by
the method of (Bolanle et al., 2015).
The agar overlay method was employed to determine
the ability of the viable lactic acid bacteria strains to
inhibit the growth of the indicator pathogens, E.coli and
Staphylococcus aureus. A loop full of LAB in MRS broth
was inoculated on MRS agar plate as a thick line of
about 2mm and about 30mm long at a good away from
the edge of the plates and incubated under microaero-
phillic condition at 37C for 24 hours. After incubation,
the MRS agar plates were overlaid with approximately
0.2ml × 10ˉ
7
CFU /ml of an overnight broth culture of the
test pathogens inoculated in 10ml of Muller Hinton soft
agar (with 0.7% agar-agar). The overlay was allowed to
set and incubated at 37C under aerobic condition. The
plates were then examined for zone of inhibition around
the line of the LAB and the clear zones were measured.
RESULTS AND DISCUSSION
A total of 56 LAB isolates were identi ed from vari-
ous freshly drawn raw milk samples collected from sur-
rounding of Tirupur and Erode district (Table 1).
All the fty six isolates were gram positive, non-
motile in hanging drop method, fty isolates were cata-
lase negative and only six showed positive to catalase
test were not a LAB. The cell morphology of fty six
isolates was evaluated through grams reaction micro-
scopic observation and majority of 36 were found to be
rods and the remaining of 20 isolates were cocci shaped.
Among the isolates 42 were able to produce CO
2
from
glucose fermentation. This result showed that they were
heterofermentative and remaining of 14 isolates were
homofermentative. Homofermentative LAB utilize glu-
cose via EMP pathway and heterofementors utilize via
HMP (hexose monophosphate pathway) described by
Rattanachaikunsopon and Phumkhachorn (2010). All the
isolates grew at 37°C but only 21 isolates were grew at
15°C;in 45°C and 55°C, only ten isolates were showed
limited growth. In 3.5 % Na Cl concentration all the iso-
lates grew well and only 27 showed growth at 6.5% Na
Cl concentration. 21 isolates showed limited growth at
18% Na Cl concentration.
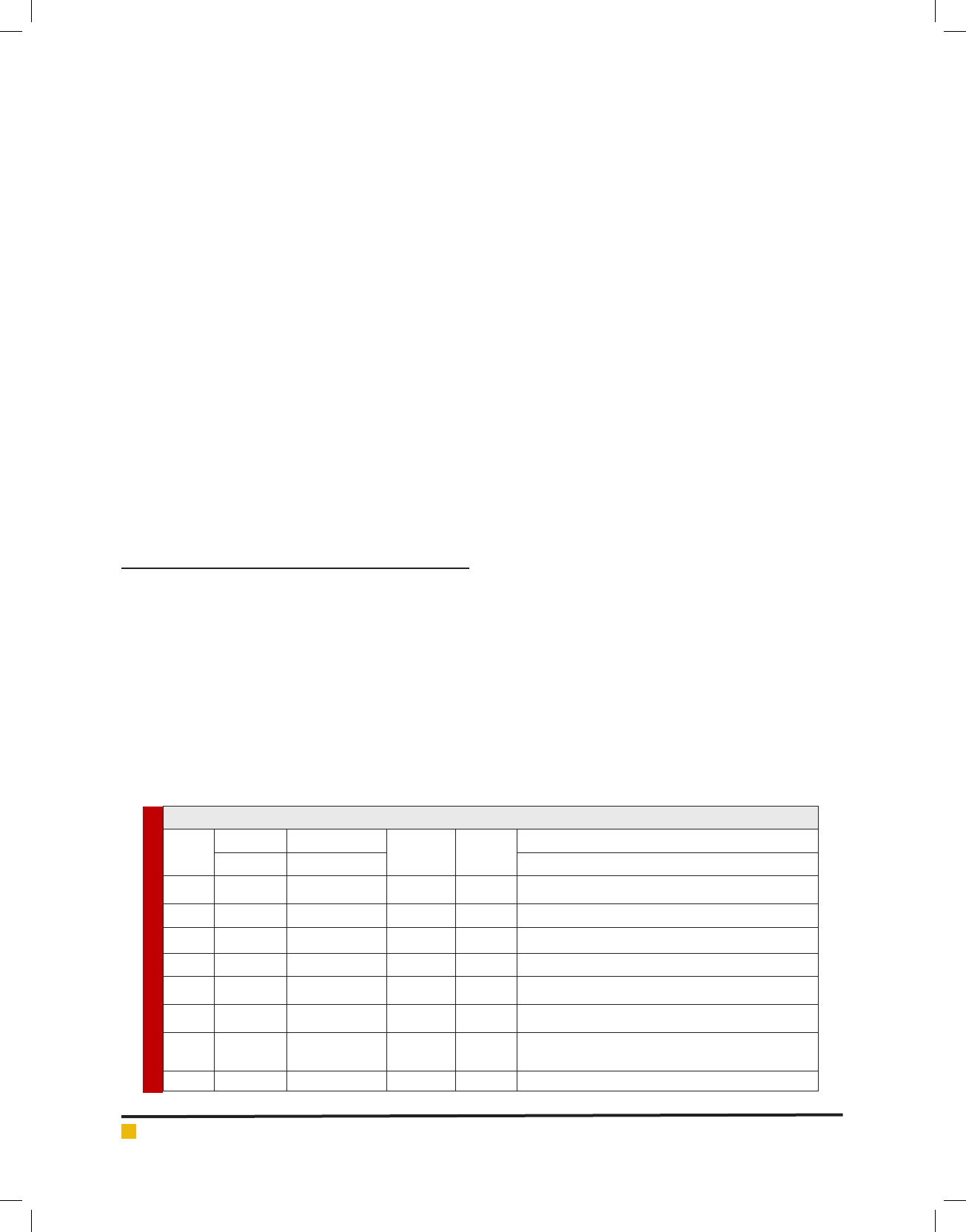
Based on the morphological, physiological and bio-
chemical test chemometric result hierarchical cluster
analysis (AHC) for nding homogenous group of iso-
lates and graphics were performed using XLStat soft-
ware version 2018.5, Addinsoft. Dendrogram showing
the similarity relationship among the 56 isolates of LAB
obtained from various raw milk. Similarities were calcu-
lated by the simple matching coef cient and grouping
was performed by Agglomeration hierarchical cluster
(AHC) analysis using Un-weighted pair group average
linkage analysis. A total of 56 isolates characterized
using set of 17 phenotypic test. An abridged dendro-
gram depicting the similarity relationship among the
isolates were divided into six groups shown in gure 2.
While the phenotypic characterization of the main clus-
ters were summarized in linkage analysis gure 1.
Results showed that all the examined strains did not
exhibited haemolysis and most of the strains were
haemolytic (non-pathogenic), while seven exhibited
haemolysis. Out of seven, ve isolates were catalase pos-
itive there were not a LAB avoided of further work.Some
of Enterococcus showed haemolysis and as well as
haemolysis summarized by W.Liu et al (2014). According
to the European Food Safety Authority (2012) antibiotics
of Tetracyclin, Streptomycin and Kanamycin were used
for antibiotic suscebtibility testing of the isolated LAB.
Table 1. Origin and designation of the isolates of Lactic acid bacteria
S. No
No. of
samples
No. of
isolates
Sample Region Designation of Strains
1 College Tirupur, Erode 2 5 C2, C9, C16, C7, C19
2 Sindhu Tirupur, Erode 11 11 C3, C5, C11, C13, C14,C17, C18, C20, C21, C22, C23
3 Cooli Tirupur, Erode 2 5 C1, C6, C8, C12, C24
4 Jersey Tirupur, Erode 1 3 C4, C10, C15
5 Buffalo Tirupur, Erode 2 5 B25, B26, B27, B28, B29
6 Goat (Black) Tirupur, Erode 4 8 G4,G15,G16,G21,G25,G26,G27,G1
7 Goat (white) Tirupur, Erode 12 17
G2,G6,G13,G14,G5,G10, G7,G8,G12,G17,G9,G18,
G19,G20,G22,G23,G24
8 Sheep Tirupur, Erode 2 2 S3,S11
428 SCREENING OF INDIGENOUS ACTIVE LACTIC ACID BACTERIA ISOLATED BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
Selvajeyanthi and Hemashenpagam
FIGURE 1. Agglomeration hierarchical clustering (AHC) of 56 isolates in raw milk
FIGURE 2. Dendrogram showed mainly six classes of 56 isolated Lactic acid bacteria in AHC
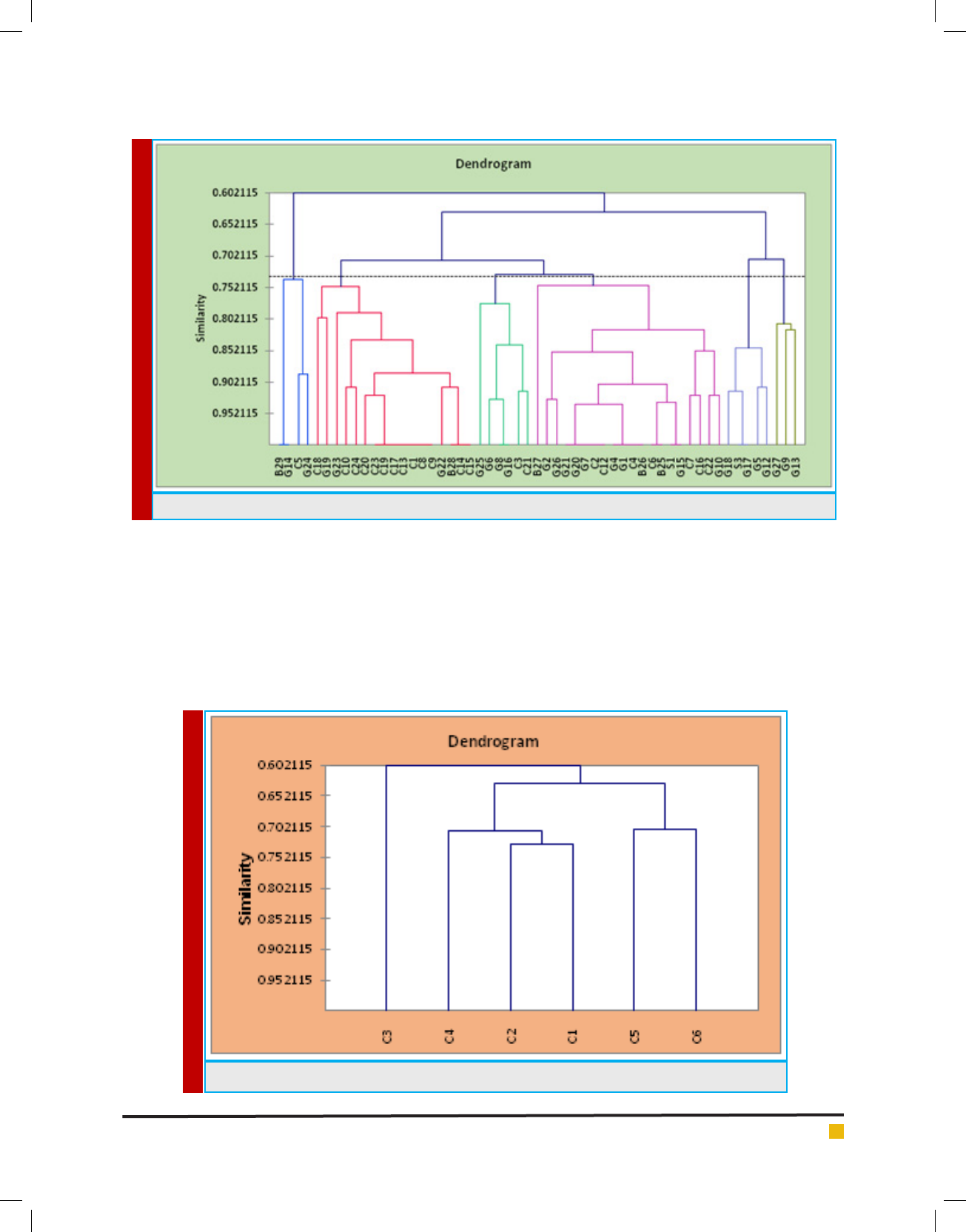
Results showed in gure 3 descripes 50% of the isolates
were resistant to antibiotics. A similar observation was
given by Hawaz (2014) who also noticed Ethiopia curd
had Lactobacillus and L. delbruekii, L.brevis and L. casei
were resistance to antibiotic streptomycin and genta-
mycin; L. fermentum, L. lactis and L. rhamnosus were
resistant to streptomycin. L. leichmanni, L. acidophillus
and L. coagulants, which were only sensitive to antibi-
otics. Furthermore Pundir et al (2013) had reported LAB
isolates from food sample were found to be resistance
to most of the antibiotics. Zarour et al (2012) had also
reported goat and camel milk isolates were resistant to
Vancomicin the strains had a poly resistance to antibiot-
ics which is attributed to plasmid transposons in many
bacterial strains. Based on the frequency of antibiotic
sensitive isolates were taken for further analysis.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS SCREENING OF INDIGENOUS ACTIVE LACTIC ACID BACTERIA ISOLATED 429
Selvajeyanthi and Hemashenpagam
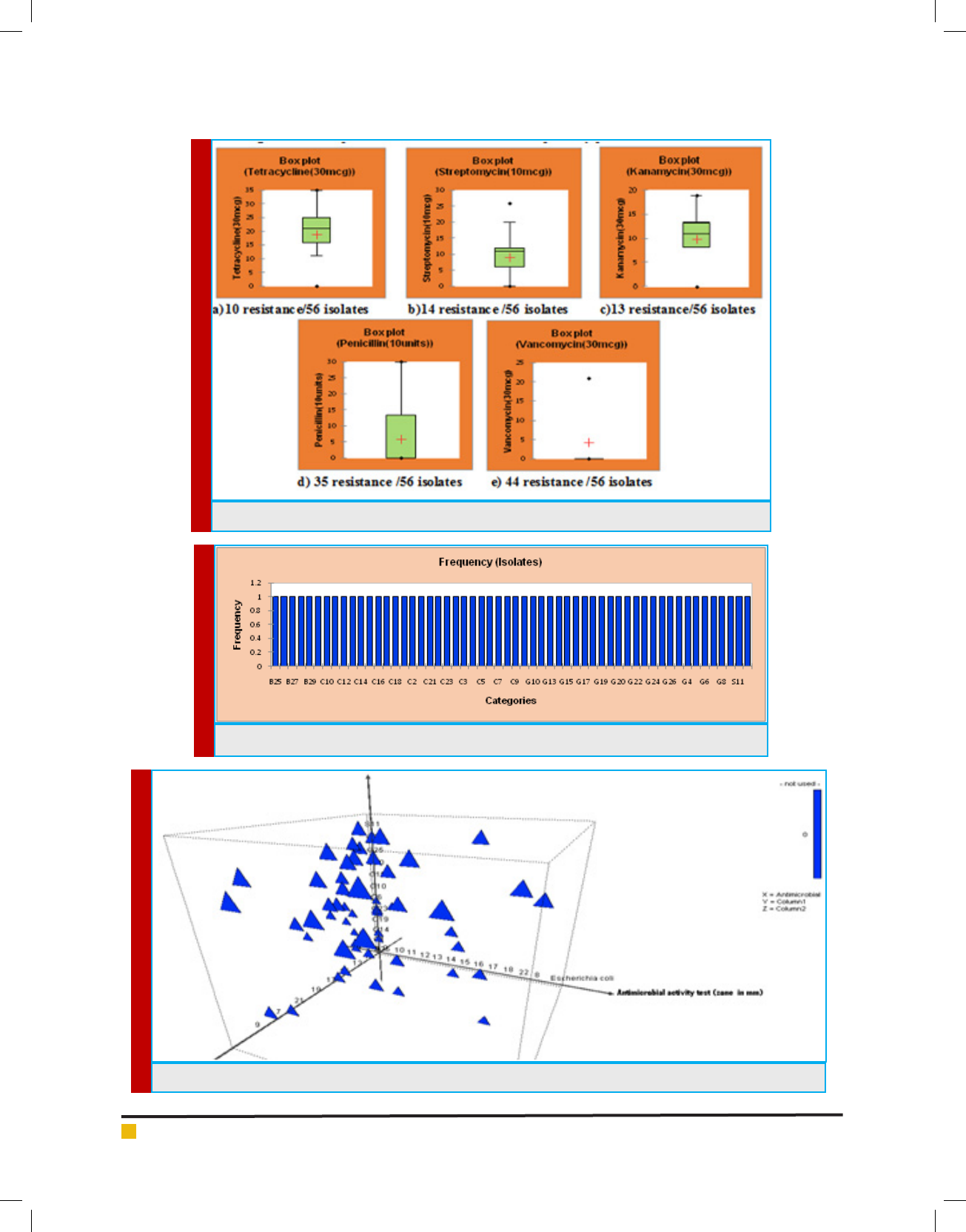
FIGURE 4. Frequency of the 28 isolates were taken for further processing based on antibiogram
FIGURE 5. Graphical 3D Scatter plot of antimicrobial activity against E.coli and S.aureus
FIGURE 3. Descriptive statistics of Antibiotic susceptibility pattern of LAB isolates
430 SCREENING OF INDIGENOUS ACTIVE LACTIC ACID BACTERIA ISOLATED BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
Selvajeyanthi and Hemashenpagam
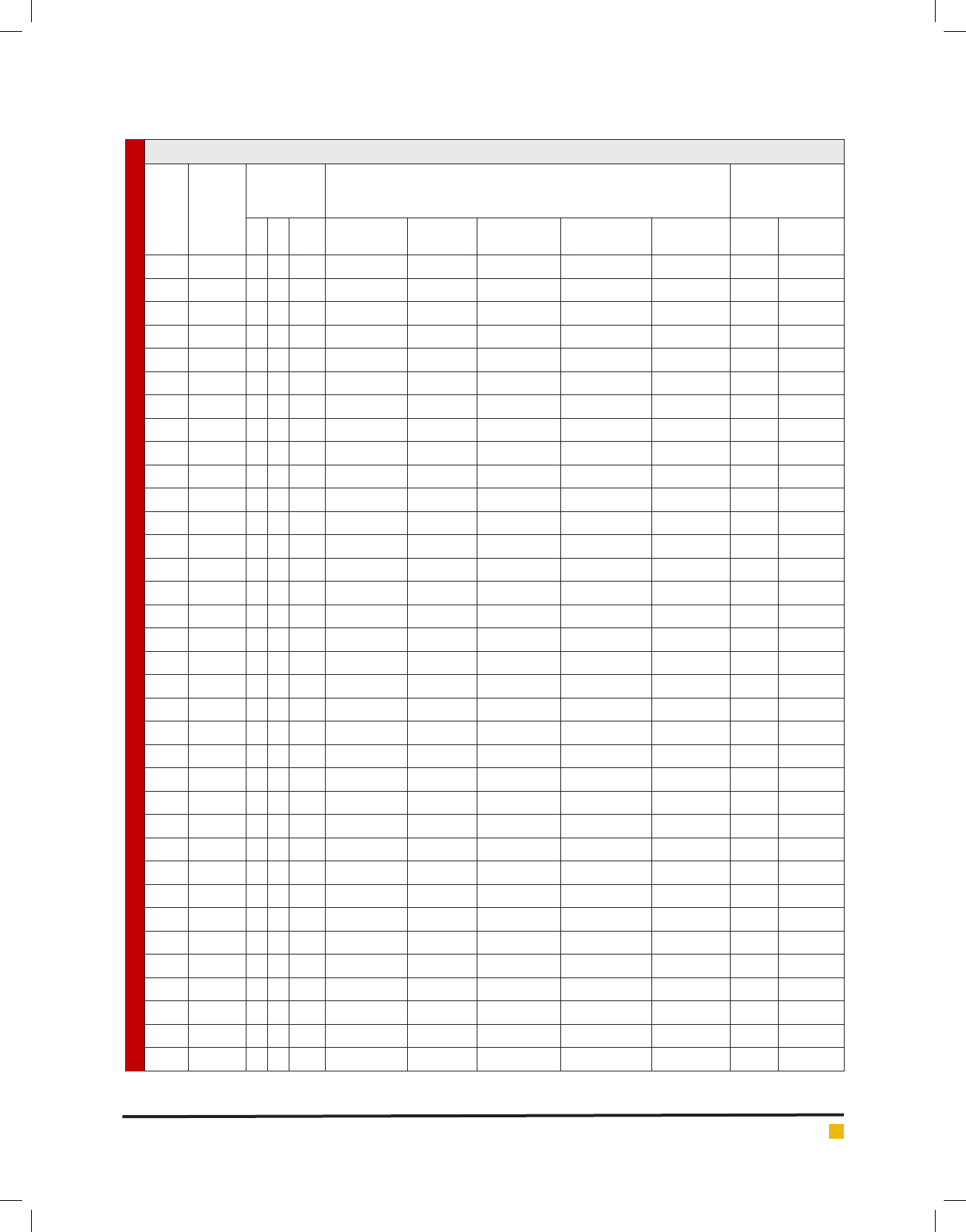
Table 2. Blood haemolysis, Antibiogram and antimicrobial activity of LAB isolates
S. No
Isolates
Blood
haemolysis
test
Antibiotic susceptibility test (zone in mm)
Antimicrobial
activity test
(zone in mm)
Tetracycline
(30 mcg)
Penicillin
(10 units)
Vancomycin
(30 mcg)
Streptomycin
(10 mcg)
Kanamycin
(30 mcg)
E. coli S. aureus
1C1 ---R R R R R 2219
2 C2 - - - 22 R R 9 14 - 13
3 C3 - - - 25 10 R 12 12 - 14
4 C4 + - - 23 R R 10 8 - 19
5C5 ---R R R R R - 13
6C6 +--15 R R 10 11 - -
7 C7 - - - 29 R R 12 11 11 11
8 C8 - - - 31 R R 13 14 - 16
9 C9 - - - 23 9 R 12 12 - 14
10 C10 - - - R R R R R 14 18
11 C11 - - - R R R R R - -
12 C12 - - - 17 13 17 12 13 16 14
13 C13 - - - 22 20 R 11 12 - 13
14 C14 - - - R R R R R - -
15 C15 - - - 31 R R 12 12 - 13
16 C16 + - - 19 R R 11 12 - 12
17 C17 - - - 22 15 R 11 11 - 12
18 C18 - - - 33 R R 12 12 18 16
19 C19 - - - 32 10 R 16 15 14 21
20 C20 - - - 27 14 R 13 11 16 13
21 C21 - - - 22 13 R 10 11 14 18
22 C22 - - - 27 12 R 11 11 15 11
23 C23 - - - 32 21 R 26 11 - -
24 C24 - - - 21 22 13 20 17 - -
25 B25 + - - 18 R 19 12 13 - 22
26 B26 + - - R R R 8 10 - 16
27 B27 + - - 23 R R 13 16 - 15
28 B28 - - - 21 R R 10 11 - 11
29 B29 - - - 31 R 21 11 12 - 8
30 G1 - - - R R R R R - 20
31 G2 - - - R R R R R - 14
32 S3 - - - 35 19 19 13 14 13 17
33 G4 - - - 22 R R 9 11 - 11
34 G5 - - - 34 14 20 15 18 10 9
35 G6 - + - 21 R R 10 8 - 12
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS SCREENING OF INDIGENOUS ACTIVE LACTIC ACID BACTERIA ISOLATED 431

Selvajeyanthi and Hemashenpagam
36 G7 - - - 21 R R 15 16 - -
37 G8 - - - 22 R R 10 10 11 19
38 G9 - - - 17 16 16 13 14 16 -
39 G10 - - - 16 R R R R - 15
40 S11 - - - 24 R R R 17 - 11
41 G12 + - - 30 14 16 11 12 - 16
42 G13 - - - 21 30 20 14 18 14 8
43 G14 - - - 30 17 R 13 13 13 15
44 G15 - - - 16 R 19 11 13 8 10
45 G16 - - - 22 R R 11 11 - 14
46 G17 - - - 18 15 20 13 17 15 8
47 G18 - - - 14 R R 11 12 12 7
48 G19 - - - 25 14 R R 19 10 -
49 G20 - - - 17 R R 12 14 17 20
50 G21 - - - 16 R R R R - 17
51 G22 - - - 24 R 20 R R - 9
52 G23 - - - R R R R R - -
53 G24 - - - 21 R R 11 10 - 13
54 G25 - - - R R R R R - 12
55 G26 + - - 11 10 R 10 11 14 22
In this study, the antimicrobial activity showed 46
isolates produced maximum inhibition zone against bac-
terial pathogen Staphylococcus sp., comparatively less
isolates of 22 produced zone against E.coli sp. Showed
in gure 4. The present work contrary with Hawaz (2014)
had reported Lactobacillus strains showed antibacterial
effect against the pathogenic E.coli, Staphylococcus sp.,
and Salmonella thyphimurium. This may vary from type
of strain and class of bacteriocin produced by LAB.
CONCLUSION
The present study concluded that freshly drawn animal
milk had rich source of lactic acid bacteria and predomi-
nant 64% Lactobacillus sp. and 34% of isolates were
coccus that tentatively had Enterococcus, Leuconostoc,
Streptococcus, Lactococcus and Pediococcus which is
in conformity with earlier work. As raw milk is used
more in Tamil Nadu being consumed frequently and
occasionally, consumers received signi cant amount of
LAB. Potentialities of probiotic characteristics are being
done in further studies. Indeed, a natural preservation of
characterized LAB in under investigation. Further work
is required on species level identi cation of isolates and
bioactive compounds produced by LAB.
ACKNOWLEDGEMENT
This work was carried out in AWECARE laboratory,
Erode TN and supported by Dept. of Microbiology Tirup-
pur Kumaran College for Women and Hindusthan Col-
lege. The support of Muthuselvi and Mehala helped in
getting samples is heart fully acknowledged.
REFERENCES
Ahmed M.M. Mabrouk, Baher A.M. Effat, Zainab I.M. Sadek,
N.F. Taw ek, Z.M.R. Hassan and Mohamed N.I. Magdoub
(2014) Antimicrobial activity of some Lactic acid bacteria Iso-
lated from Egyptian Dairy Products, International Journal of
Chem Tech Research CODEN (USA) Vol.6, No.2, pp 1139-1150
Ashwani Kumar Singh, Abhishek Pandey, Megha Sharma,
Komudi and Anupam Singh (2014) “Probiotic activities of lac-
tic acid bacteria isolated from human breast milk, Journal of
Biological Engineering Research and Review., Vol.1, Issue 2,
07-12, www.biologicalengineering.in
Ayman Ashmaig, Alaa Hasan and Eisa El Gaali (2009) Identi-
cation of lactic acid bacteria Isolated from traditional Suda-
nese fermented camel’s milk, African journal of Microbiology
Research Vol.3 (8) pp. 451-457 http://www.academicjournals.
org/ajmr
Bolanle A. Adeniyi, Adewale Adetoye, Funmilola A (2015)
Ayeni Antimicrobial activities of lactic acid bacteria isolated
432 SCREENING OF INDIGENOUS ACTIVE LACTIC ACID BACTERIA ISOLATED BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS

Selvajeyanthi and Hemashenpagam
from cow faeces against potential enteric pathogens, African
health sciences, Vol 15 issue 3: 888-895.
Delavenne. E, J. Mounier, F. Déniel, G. Barbier and G. Le Blay
(2012) Biodiversity of Antifungal Lactic Acid Bacteria Isolated
from Raw Milk Samples from Cow, Ewe and Goat over One-
Year Period, International Journal of Food Microbiology, Vol.
155, No. 3, pp. 185-190.
European Food Saftey Authority (EFSA) – committed since
2002 to ensuring that Europe’s food is safe: http://www.efsa.
europa.eu/ en/search.htm?text=probiotics8p=10]
Geeta Shukla, Gatha Sharma and Nisha Goyal (2010) Probiotic
characterization of Lactobacilli and yeast strains isolated from
Whey beverage and Therapeutic potential of Lactobacillus
yoghurt in Murine Giardiasis, American journal of biomedical
sciences vol-2(3): 248-261.
Hawaz E (2014) Isolation and identi cation of probiotic lactic
acid bacteria from curd and in vitro evaluation of its growth
inhibition activities against pathogenic bacteria, African
Journal of Microbiology Research vol. 8 (13), pp. 1419-1425,
DOI:10.5897/AJMR14.6639 http://www.academicjournals.org/
AJMR, Article No. D80ED0B46925
Kailasapathy K, Phillips H M (2008) Survival of lactobacillus aci-
dophilus and Bi dobacterium animalis sp., lactis is stirred fruit
yougurts. LWT-Food Science Technology; Vol. 41,1317-1322
Liu, W H. Zhang and H. Pang, Y. Cai, Lactic acid bacteria,
Chapter 2 Biodiversity of Lactic acid bacteria, ©Springer Sci-
ence + Business Media Dordrecht 2014, DOI:10.1007/978-94-
0178841-0
-
2
Marakoudakis P.A.M Papadelli, M Georgalaki, E G Panayoto-
poulou, B M Gonzalez, A.F. Mentis, K. Petraki, DS Sgouras and
E Tsakalidou (2009) In vitro and in vivo safety evaluation of
bacteria in producer, Streptococcus macedonicus ACA-DC198.
International Journal of Food Microbiology, 133:141-147
Mduduzi Paul Mokoena,Taurai Mutanda and Ademola O.
Olaniran (2016) “Review Article-Prospectives on the probi-
otic potential of lactic acid bacteria from African traditional
fermented foods and beverages, Food and Nutrition research,
60;29630-http//dx.doi.org/10.3402fnnv60.29630
Oktay Yerlikaya (2014) Starter cultures used in probiotic dairy
drinks, Food Science and Technology, campinas, 34(2):221-229
DOI:http://dx.doi.org/10.1590/fst2014-0050
Pooja Thakkar, H.A. Modi and J.B. Prajapati (2015) “Isolation,
characterization and safety assessment of lactic acid bacteria
isolate from fermented food products,” Int. J. curr. Microbiol.
App. Sci; 4(4); 713-725
Rattanachaikunsopon P and P. Phumkhachorn (2010) “Lactic
acid bacteria: their antimicrobial compounds and their uses in
food production,” Annals of Biological Research,1(4): 218-228
www.scholarsresearchlibrary.com
Ram Kumar Pundir, Satish Rana, Neha Kashyap and Amandeep
kaur (2013) Probiotic potential of lactic acid bacteria isolated
from food samples:an in vitro study, Journal of Applied Phar-
maceutical Science vol. 3 (03), pp. 085-093, DOI:10.7324/
JAPS.2013.30317 http://www.japsonline.com
Subhashini (2014) Bioprospecting of LAB for potentiality as
probiotics International Journal of Microbiological Research
5(2); 90-97 DOIL:10.5829/idosi.ijmr.2014.5.2.83291
Setyawardani T et al. (2011) Potential probiotic isolates of Lac-
tobacillus rhamnosus and L.plantarum were present in indige-
nous goat milk, Animal Production13(1):55-63 http://www.
researchgate.net/publication/228472966
Wassie Misganaw and Wassie Teketay (2016) Isolation and
identi cation of Lactic acid bacteria from Raw cow milk, Inter-
national journal of Advanced research in Biological Sciences
vol. 3(8) pp. 44-49
Wouters J.T, Ayad E.H, Hugenholtz J, Smit G (2002) Microbes
from raw Milk for Fermented Dairy Products, International
Dairy Journal: Vol.12 No.2-3., pp.91-109
Zarour K, Benmechernene Z, Hadadji M,Moussa-Boudjemaa
B, Henni D.J and Kihal M (2012) Bioprospecting of Leuconos-
toc mesenteroids strains isolated from Algerian raw camel and
goat milk for technological properties useful as adjunct start-
ers, African Journal of Microbiology Research vol. 6(13), pp.
3192-3201, DOI:10.5897/AJMR11.1542 http://www.academic-
journals.org/AJMR
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS SCREENING OF INDIGENOUS ACTIVE LACTIC ACID BACTERIA ISOLATED 433