1*Postgraduate Department Of Zoology, Raja Narendra Lal Khan Women’s College (Autonomous) (Natural And
Applied Science Research Centre), Paschim Medinipur, 721102, West Bengal, India
2 Department Of Zoology, Dr. Bhupendranath Dutta Smriti Mahavidyalaya, Hatgobindapur, 713407, West Bengal, India
3department Of Zoology, Vidyasagar University, Paschim Medinipur, 721102, West Bengal, India
Corresponding author email: ppcrnlk@gmail.com
Article Publishing History
Received: 03/11/2020
Accepted After Revision: 24/12/2020
Bacterial populations from organs (viz., liver, spleen, kidney and gill) of Clarias batrachus of the sewage fed water areas near IISCo slag disposal site, Dharampur on northern side of Damodar River, Asansol, West Bengal, India, were enumerated, followed by determination of resistance for antibiotics and heavy metals. Maximum resistance is shown against ampicillin (95%) and minimum against ciprofloxacin (5%). Most of the isolates exhibited an increasing order of tolerance for the metals (µg/mL) copper (200), cadmium (200), iron (400) and chromium (400), with minimum inhibitory concentration (MIC) ranging from <50 to 1600 µg/mL. A total of 100 bacteria have been successfully isolated from internal organs of of Clarias batrachus (Aeromonas species (20%); Escherichia coli, (45%); Bacillus species (4%); Pseudomonas aeruginosa (6%), Staphylococcus aureus (18%) and coagulate-negative Staphylococci aureus (7%)). In terms of antibiotic susceptibility testing, each isolate was tested against 10 antibiotics. The multiple antibiotic resistance (MAR) index the isolated bacterial ranged fro 0.2-0.7. These observations indicate that the bacteria isolates are from a high risk source where antibiotics are frequently used, possibly from sewage effluents. Significant occurrence of bacterial population in organs of fish with high incidence of resistance for antibiotics and heavy metals may pose risk to fish fauna and public health.
Clarias batrachus, Antibiogram, Heavy Metal Resistance, Multiple Antibiotic Resistance
Deb A, Bhattacharjee I, Das T, Mandal B, Chakravorty P. P. Antibiotic and Heavy Metal Resistance in Bacteria from Organs of Sewage Fed Farm Fishes. Biosc.Biotech.Res.Comm. 2020;13(4).
Deb A, Bhattacharjee I, Das T, Mandal B, Chakravorty P. P. Antibiotic and Heavy Metal Resistance in Bacteria from Organs of Sewage Fed Farm Fishes. Biosc.Biotech.Res.Comm. 2020;13(4). Available from: <a href=”https://bit.ly/2Jg4WBw”>https://bit.ly/2Jg4WBw</a>
Copyright © Deb et al., This is an Open Access Article distributed under the Terms of the Creative Commons Attribution License (CC-BY) https://creativecommons.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
Clarias batrachus, commonly called asian catfish is a threatened (Hossain et al., 2006; Ahmad et al., 2012; Roy et al., 2018), and critically endangered species (Binoy, 2010). It‘s a promising hardy fish, excellent nutritional profile and market price is high (Hossain et al., 2006; Goswami 2007; Debnath 2011).Tham et al., (2009) have reported heavy metal inhibitions by AchE from C. batrachus. Heavy metals are ubiquitous and persist as environmental pollutants that are introduced into the environment through anthropogenic activities, like mining and smelting, also well as through irrigation and other sources of commercial waste. However, untreated or partially treated wastewaters introduce an enormous amount of contaminates particularly heavy metals into agricultural lands (Wang and Tao, 1998; Boateng et al., 2019). The existence of heavy metals within the environment represents a big and long-term environmental hazard since they’re not biodegradable and have a tendency to accumulate in living organisms (Kobya et al., 2005; Liao et al., 2008; Genchi et al., 2020).
Indiscriminate use of various antibiotics has caused development of resistance for various antimicrobials and chemotherapeutic agents among the gut flora of homeotherms. Use of antibiotics will exert more selective pressure and resistant pathogens are going to be encontered more frequently (MacMillan, 2001; Priyadarshini et al., 2020). Resistance to antibiotics and metals occurs simultaneously when the genes specifying resistant phenotypes are located together an equivalent genetic element like a plasmid, transposon, or integron (Chapman 2003; Frost et al., 2005; Venner et al., 2009; McMillan et al. 2019). This present study was conducted to evaluate the antibiotic and heavy metal tolerance of bacteria obtained from internal organs of Clarias batrachus.
MATERIAL AND METHODS
Clarias batrachus were collected from the sewage fed water areas near IISCo slag disposal site, Dharampur on northern side of Damodar River at an altitude of 75 meters. Longitudinally it is at 86°55ʹ East and latitudinally at 23°40ʹ North Asansol, West Bengal, India.Total of 180 fish samples were collected from June to August 2019. 60 samples were collected at the first visit and was repeated three times. Fish were collected in sterile plastic bags with labelling from sites. Fish samples were transported to the laboratory in cooler box and tested on the same day.
Fish samples were sacrificed to dissect out aseptically the liver, spleen, kidney and gills (Pathak and Gopal, 2005) placed in labelled bottle containing peptone water and buffered peptone water. The contents were homogenized and the homogenate from peptone water was streaked using a sterile loop on blood agar and MacConkey agar. Streaked plates were incubated at temperature (37 °C) aerobically for 24 hours. Homogenate in buffered peptone water was incubated for 24 hours at 37 °C. After incubation, 1 ml of the homogenate tranferred into Rappaport Vassiliadis (RV) agar and icubated for 24 hours at 37 °C. On third day, loopful of the sampole containing bacteria was streaked on Xylose Lysine Deoxychocolate (XLD) agar and incubated for 24 hours at 37 °C (Markey et al. 2013).
The isolated bacteria were identified using morphological characteristics, Gram staining and biochemical tests (oxidase, motility, indole, citrate, lysine decarboxylase, urease. Triple sugar iron). The scheme of Cowan and Steel (1993) was followed for characterization and identification of strains, and the resulkts were interpreted using Bergey’s Manual of Systematic Bacteriology (Staley et al. 1989). Autoclaved fish and uninoculated media were used as negative controls. Different strains of bacteria obtained from MTCC Chandiagrh, India (Aeromonoas 646, Bacillus niacini MTCC 8323, Escherichia coli MTCC 739, Pseudomonas aeruginosa MTCC 2453, Staphylocccus aureus MTCC 2940) were used as positive control.
Antibiotic susceptibility testing was conducted consistent with Kirby and Bauer disk diffusion method (Bauer et al., 1966) by employing commonly used antibiotics (namely 30 µg of amikacin, 30 µg of amoxicillin, 10 µg of ampicillin, 30 µg of chloramphenicol, 5 µg of ciprofloxacin, 10 µg of gentamycin, 5 µg of levofloxacin, 10 µg of sparfloxacin, 25 µg of streptomycin and 30 µg of tetracyline). The colonies were transferred into agar plate. The swab was then streaked in three, different directions over the surface of plate of Mueller-Hinton Agar such that a uniform well spread out inoculum is achieved. After 18 hours of incubation at a specific temperature (37±1 °C) the plates were examined and the diameters of the inhibition zone was measured to the nearest millimeter. Inhibitions were measured and the result interpreted using Clinical Laboratory Standard Institute (2017).
The strain isolated was tested to four metals by Agar dilution method (Malik and Ahemad, 2006; Kinare and Shingadia, 2014). Stock solutions of 104 μg/mL were prepared by dissolving the precise quantities of the follwing metal salts: CdCl2 (SRL), K2Cr2O7 (SIGMA-ALDRICH), CuSO4. 5H2O(SIGMA-ALDRICH) and FeCl3 (MERCK) in water and sterilized. 20 mL of agar was poured into petri plates and therefore the volume of metal stock solutions was calculated by the formula: C1 × V1 = C2 × V2, where C1 is the metal concentration avaiable, V1 is the volume of stock solution used, C2 is that the concentration of metal in agar, and V2 is that the volume of agar. Then the isolated strains were streaked onto the medium-containing increasing concentrations of metal salts using sterile loops. Then, plates were sealed and incubated at 30 °C for five days. Plate-containing only agar was also inoculated and incubated to act as control. Rock bottom concentration of every metal at which no growth occurred in comparison to the control plates was considered as MIC value.
The multiple antibiotic resistance (MAR) index of isolates against tested antibiotics were determined as described by Krumperman (1983) using the formula: a/b, where “a” represents the number of antibiotics against which a particular isolate was resistant and “b” the total number of antibiotics used for test.
RESULTS AND DISCUSSION
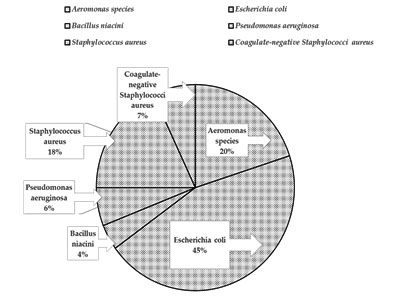
The bacteria isolated were Aeromonas species, Escherichia coli, Bacillus niacini, Pseudomonas aeruginosa, Staphylococcus aureus, and coagulate-negative Staphylococci aureus which were identified using morphological properties, gram staining, and series of biochemical tests. Total 100 bacteria were isolated from liver, spleen, kidney and gill. The bacterial load, i.e., total viable count, was found to be 5.62 × 104, 4.12 × 104, 2.30 × 104 and 1.76 × 104 c.f.u./mL in liver, spleen, kidney and gill of the experimental fish, respectively. The percentage prevalance of the isolated bacteria were Aeromonas species (20%); Escherichia coli, (45%); Bacillus species (4%); Pseudomonas aeruginosa (6%), Staphylococcus aureus (18%) and coagulate-negative Staphylococci aureus (7%). Prevalence rate of isolated bacteria are plotted in (Figure 1).
The antibiotic resistance among random bacterial isolates from all four organs has shown a full range of resistance (0–100%) for ten common antibiotics of therapeutic and prophylactic uses among human beings, and in fish aquaculture. Resistance was found to be maximum among the isolates from spleen, kidney, and liver, while it was minimum among those from gill. Maximum average resistance was exhibited for ampicillin (95%) and tetracycline (75%) and minimum for ciprofloxacin (05%) (Table 1).
The heavy metal resistance percentage among bacterial isolates from fish organs presented in Table 2. The maximum tolerance, in general, was observed for chromium and iron (400 µg/mL), while it was minimum for copper and cadmium (200 µg/mL). The MIC value of isolates is presented in Table 3. Test isolates were also found to be tolerant to different concentrations of various toxic heavy metals as evidenced by their MICs ranging from <50 to 1600 µg/mL (Table 3).
Except some gill isolated bacteria (ADCG14-ADCG21) the MAR index of all isolates show values higher than 0.2 (Table 4). All the isolate (except ADCG14-ADCG21) were multidrug resistant (resistant to three or more drugs). The trend of MAR index is alarming for the bacterial species isolated from the site.
Figure 1: Prevalance rate of the isolated bacteria
Table 1. Antibiotic resistance (%) among bacterial isolates from fish organs
| Antibiotics (µg/mL) | Fish organs | Average resistance | ||||
| Liver | Spleen | Kidney | Gill | |||
| Amikacin (AMK) (30) | 90 | 80 | 60 | 30 | 65 | |
| Amoxycillin (AMX) (25) | 90 | 80 | 70 | 40 | 70 | |
| Ampicillin (AMP) (10) | 100 | 100 | 90 | 90 | 95 | |
| Chloramphenicol (CHL) (30) | 80 | 50 | 50 | 20 | 50 | |
| Ciprofloxacin (CIP) (5) | 10 | 10 | 00 | 00 | 05 | |
| Gentamycin (GEN) (10) | 50 | 40 | 20 | 10 | 30 | |
| Levofloxacin (LVX) (5) | 50 | 40 | 10 | 00 | 25 | |
| Sparfloxacin (SPX) (10) | 60 | 50 | 30 | 20 | 40 | |
| Streptomycin (STR) (25) | 20 | 10 | 10 | 00 | 10 | |
| Tetracyline (TET) (30) | 100 | 80 | 70 | 50 | 75 | |
Table 2. Heavy metal resistance (%) among bacterial isolates from fish organs
| Heavy metals (µg/mL) | Fish organs | Average resistance | |||
| Liver | Spleen | Kidney | Gill | ||
| Copper (200) | 10 | 00 | 00 | 00 | 2.5 |
| Chromium(400) | 100 | 100 | 80 | 70 | 87.5 |
| Cadmium (200) | 30 | 20 | 10 | 00 | 15 |
| Iron (400) | 90 | 100 | 80 | 50 | 80 |
Table 3. MIC values for different heavy metals among bacterial isolates from fish organs
| Heavy metals | MIC values (µg/mL) for different Fish organs | |||
| Liver | Spleen | Kidney | Gill | |
| Copper | < 50 | – | – | – |
| Chromium | 1600 | 1600 | 800 | 400 |
| Cadmium | 100 | 50 | < 50 | – |
| Iron | 1600 | 800 | 800 | 400 |
Table 4. Code number assigned with organs from which the bacteria is isolated, antibiotic resistance Profile and multiple antibiotic resistance index of the isolated bacteria from site
| Sl No | Code No assigned to isolate | Organs from Isolated | Antibiotic resistant Profile | Resistant to number of Antibiotics | MAR Index |
| 1 | ADCL1 | Liver | AMK,AMX,AMP,TET,CHL,GEN | 6 | 0.6 |
| 2 | ADCL2 | Liver | AMK,AMX,AMP,TET, CHL,LVX | 6 | 0.6 |
| 3 | ADCL3 | Liver | AMK,AMX,AMP,TET, CHL,GEN,SPX | 7 | 0.7 |
| 4 | ADCL4 | Liver | AMK,AMX,AMP,TET, CHL,LVX,SPX | 7 | 0.7 |
| 5 | ADCL5 | Liver | AMK,AMX,AMP,TET, CHL,GEN,SPX | 7 | 0.7 |
| 6 | ADCL6 | Liver | AMK,AMX,AMP,TET, CHL,LVX,SPX | 7 | 0.7 |
| 7 | ADCL7 | Liver | AMK,AMX,AMP,TET, CHL,GEN,SPX | 7 | 0.7 |
| 8 | ADCL8 | Liver | AMK,AMX,AMP,TET, CHL,LVX,SPX | 7 | 0.7 |
| 9 | ADCL9 | Liver | AMK,AMX,AMP,TET, CHL,GEN,SPX | 7 | 0.7 |
| 10 | ADCL10 | Liver | AMK,AMX,AMP,TET, CHL,LVX,SPX | 7 | 0.7 |
| 11 | ADCL11 | Liver | AMK,AMX,AMP,TET, CHL,GEN,SPX | 7 | 0.7 |
| 12 | ADCL12 | Liver | AMK,AMX,AMP,TET, CHL,GEN,SPX | 7 | 0.7 |
| 13 | ADCL13 | Liver | AMK,AMX,AMP,TET, CHL,LVX,SPX | 7 | 0.7 |
| 14 | ADCL14 | Liver | AMK,AMX,AMP,TET, CHL,LVX,SPX | 7 | 0.7 |
| 15 | ADCL15 | Liver | AMK,AMX,AMP,TET, CHL,LVX,SPX | 7 | 0.7 |
| 16 | ADCL16 | Liver | AMK,AMX,AMP,TET, CHL,LVX | 6 | 0.6 |
| 17 | ADCL17 | Liver | AMK,AMX,AMP,TET, CHL,LVX | 6 | 0.6 |
| 18 | ADCL18 | Liver | AMK,AMX,AMP,TET, CHL,GEN | 6 | 0.6 |
| 19 | ADCL19 | Liver | AMK,AMX,AMP,TET, CHL,GEN | 6 | 0.6 |
| 20 | ADCL20 | Liver | AMK,AMX,AMP,TET, CHL,GEN | 6 | 0.6 |
| 21 | ADCL21 | Liver | AMK,AMX,AMP,TET, CIP,GEN,STR | 7 | 0.7 |
| 22 | ADCL22 | Liver | AMK,AMX,AMP,TET, CIP,STR | 6 | 0.6 |
| 23 | ADCL23 | Liver | AMK,AMX,AMP,TET,CIP,LVX,STR | 7 | 0.7 |
| 24 | ADCL24 | Liver | AMP,TET,LEV,GEN,STR,SPX | 6 | 0.6 |
| 25 | ADCL25 | Liver | AMP,TET,LEV,GEN,STR,SPX | 6 | 0.6 |
| 26 | ADCS1 | Spleen | AMP,AMK,AMX,TET,GEN,LVX | 6 | 0.6 |
| 27 | ADCS2 | Spleen | AMP,AMK,AMX,TET, CHL,SPX | 6 | 0.6 |
| 28 | ADCS3 | Spleen | AMP,AMK,AMX,TET, GEN,LVX | 6 | 0.6 |
| 29 | ADCS4 | Spleen | AMP,AMK,AMX,TET, CHL,SPX | 6 | 0.6 |
| 30 | ADCS5 | Spleen | AMP,AMK,AMX,TET, CHL,SPX | 6 | 0.6 |
| 31 | ADCS6 | Spleen | AMP,AMK,AMX,TET, GEN,LVX | 6 | 0.6 |
| 32 | ADCS7 | Spleen | AMP,AMK,AMX,TET, CHL,SPX | 6 | 0.6 |
| 33 | ADCS8 | Spleen | AMP,AMK,AMX,TET, GEN,LVX | 6 | 0.6 |
| 34 | ADCS9 | Spleen | AMP,AMK,AMX,TET, CHL,SPX | 6 | 0.6 |
| 35 | ADCS10 | Spleen | AMP,AMK,AMX,TET, GEN,LVX | 6 | 0.6 |
| 36 | ADCS11 | Spleen | AMP,AMK,AMX,TET, CHL,SPX | 6 | 0.6 |
| 37 | ADCS12 | Spleen | AMP,AMK,AMX,TET | 4 | 0.4 |
| 38 | ADCS13 | Spleen | AMP,AMK,AMX,TET | 4 | 0.4 |
| 39 | ADCS14 | Spleen | AMP,AMK,AMX,TET | 4 | 0.4 |
| 40 | ADCS15 | Spleen | AMP,AMK,AMX,TET, CHL,SPX | 6 | 0.6 |
| 41 | ADCS16 | Spleen | AMP,AMK,AMX,TET, GEN,LVX | 6 | 0.6 |
| 42 | ADCS17 | Spleen | AMP,AMK,AMX,TET, CIP,STR | 6 | 0.6 |
| 43 | ADCS18 | Spleen | AMP,AMK,AMX,TET | 4 | 0.4 |
| 44 | ADCS19 | Spleen | AMP,AMK,AMX,TET,CHL,SPX | 6 | 0.6 |
| 45 | ADCS20 | Spleen | AMP,AMK,AMX,TET, CIP,STR | 6 | 0.6 |
| 46 | ADCS21 | Spleen | AMP,CHL,SPX,CIP,STR | 5 | 0.5 |
| 47 | ADCS22 | Spleen | AMP, CHL,SPX, GEN,LVX | 5 | 0.5 |
| 48 | ADCS23 | Spleen | AMP, CHL,SPX, GEN,LVX | 5 | 0.5 |
| 49 | ADCS24 | Spleen | AMP, CHL,SPX, GEN,LVX | 5 | 0.5 |
| 50 | ADCS25 | Spleen | AMP, CHL,SPX, GEN,LVX | 5 | 0.5 |
| 51 | ADCK1 | Kidney | AMK,AMX, AMP,TET | 4 | 0.4 |
| 52 | ADCK2 | Kidney | AMK,AMX, AMP,TET | 4 | 0.4 |
| 53 | ADCK3 | Kidney | AMP, CHL, SPX,TET | 4 | 0.4 |
| 54 | ADCK4 | Kidney | AMK,AMX, AMP,TET | 4 | 0.4 |
| 55 | ADCK5 | Kidney | AMP, CHL, LVX,TET | 4 | 0.4 |
| 56 | ADCK6 | Kidney | AMK,AMX, AMP,TET | 4 | 0.4 |
| 57 | ADCK7 | Kidney | AMK,AMX, LVX,TET | 4 | 0.4 |
| 58 | ADCK8 | Kidney | AMK,AMX, LVX,TET | 4 | 0.4 |
| 59 | ADCK9 | Kidney | AMX, AMP, GEN,TET | 4 | 0.4 |
| 60 | ADCK10 | Kidney | AMK AMP, GEN,TET | 4 | 0.4 |
| 61 | ADCK11 | Kidney | AMK,AMX, GEN,TET | 4 | 0.4 |
| 62 | ADCK12 | Kidney | AMX, AMP, GEN,TET | 4 | 0.4 |
| 63 | ADCK13 | Kidney | AMK,AMX, AMP,TET | 4 | 0.4 |
| 64 | ADCK14 | Kidney | AMK AMP, CHL, SPX | 4 | 0.4 |
| 65 | ADCK15 | Kidney | AMK, AMP, CHL,TET | 4 | 0.4 |
| 66 | ADCK16 | Kidney | AMX, AMP, CHL,TET | 4 | 0.4 |
| 67 | ADCK17 | Kidney | AMX, AMP, CHL,STR | 4 | 0.4 |
| 68 | ADCK18 | Kidney | AMX, AMP, CHL,SPX,TET | 5 | 0.5 |
| 69 | ADCK19 | Kidney | AMP, AMP, GEN,STR | 4 | 0.4 |
| 70 | ADCK20 | Kidney | AMK, AMP, CHL, STR | 4 | 0.4 |
| 71 | ADCK21 | Kidney | AMK, AMP, CHL, SPX,TET | 5 | 0.5 |
| 72 | ADCK22 | Kidney | AMX, AMP, CHL, SPX | 4 | 0.4 |
| 73 | ADCK23 | Kidney | AMK,AMP, CHL, SPX | 4 | 0.4 |
| 74 | ADCK24 | Kidney | AMX, AMP, CHL, SPX,TET | 5 | 0.5 |
| 75 | ADCK25 | Kidney | AMK, AMP, CHL, SPX | 4 | 0.4 |
| 76 | ADCG1 | Gill | AMK, AMP,TET | 3 | 0.3 |
| 77 | ADCG2 | Gill | AMX, AMP, TET | 3 | 0.3 |
| 78 | ADCG3 | Gill | AMK, AMP, TET | 3 | 0.3 |
| 79 | ADCG4 | Gill | AMX, AMP, TET | 3 | 0.3 |
| 80 | ADCG5 | Gill | AMK, AMP, TET | 3 | 0.3 |
| 81 | ADCG6 | Gill | AMX, AMP, TET | 3 | 0.3 |
| 82 | ADCG7 | Gill | AMX, AMP, TET | 3 | 0.3 |
| 83 | ADCG8 | Gill | AMK, AMP, TET | 3 | 0.3 |
| 84 | ADCG9 | Gill | AMX, AMP, TET | 3 | 0.3 |
| 85 | ADCG10 | Gill | AMX, AMP, TET | 3 | 0.3 |
| 86 | ADCG11 | Gill | AMK, AMP, TET | 3 | 0.3 |
| 87 | ADCG12 | Gill | AMX, AMP, TET | 3 | 0.3 |
| 88 | ADCG13 | Gill | AMK, AMP, TET | 3 | 0.3 |
| 89 | ADCG14 | Gill | AMK, AMP | 2 | 0.2 |
| 90 | ADCG15 | Gill | AMX, AMP | 2 | 0.2 |
| 91 | ADCG16 | Gill | AMX, GEN | 2 | 0.2 |
| 92 | ADCG17 | Gill | AMX, GEN | 2 | 0.2 |
| 93 | ADCG18 | Gill | AMP, GEN | 2 | 0.2 |
| 94 | ADCG19 | Gill | AMP, SPX | 2 | 0.2 |
| 95 | ADCG20 | Gill | AMP, CHL | 2 | 0.2 |
| 96 | ADCG21 | Gill | AMP, CHL | 2 | 0.2 |
| 97 | ADCG22 | Gill | AMP, CHL, SPX | 3 | 0.3 |
| 98 | ADCG23 | Gill | AMP, CHL, SPX | 3 | 0.3 |
| 99 | ADCG24 | Gill | AMP, CHL, SPX | 3 | 0.3 |
| 100 | ADCG25 | Gill | AMK, AMP, SPX | 3 | 0.3 |
Contamination of river water with municipal sewage and industrial effluent results in the occurrence of pathogenic microorganisms, particularly fecal bacteria and toxic metals, above their maximum permissible limits (Chatterjee et al., 2010, Iloms et al., 2020). Fish in such water are exposed to these bacteria and metals, which bioconcentrate in different organs of fish. It has been observed to be maximum in liver and minimum in gills. Thus, it appears from these findings that soft tissues in massive organs are more prone to bioconcentration of bacteria, leading to incidence of infectious diseases among the aquatic fauna. This may be due to availability of more nutrients and lack of exposure to the surroundings. Bioconcentration of aquatic bacteria such as coliforms, streptococci, and aeromonads in gut, liver, and muscles of tilapia fish grown in a sewage-contaminated pond has also been noticed (Fattal et al., 1993; Wamala et al., 2018).
The resistance exhibited for ciprofloxacin, levofloxacin, streptomycin and gentamycin is a signal of the effectiveness of broad-spectrum antibiotics of the present generation. With these observations it appears that the source of the problem of antibiotic resistance in riverine ecosystems is fecally contaminated water, and fish populations in them plays important role in creating resistance. Antibiotic resistance patterns in the bacterial population in an aquatic ecosystem have been found to be useful in identifying non point sources of fecal pollution (Wiggins et al., 1999; Labrador et al., 2019).
The occurrence of resistance for common antibiotics is, further, an indication of indiscriminate use of these antibiotics, leading to constraint in antimicrobial therapy for infectious diseases. The loss of antibiotic susceptibility among the aquatic bacteria has been observed to be affected to a considerable extent by the physicochemical qualities of water and seasonal variations (Pathak et al., 1993). In addition to assessment of loss of antibiotic susceptibility, the test isolates were also found to be tolerant to different concentrations of various toxic heavy metals as evidenced by their MICs. The isolates from visceral organs, i.e., spleen, kidney, and liver, exhibited maximum resistance for ampicillin, tetracycline, and amoxicillin with the highest tolerance for iron and chromium; while the isolates from gills showed minimum resistance for ampicillin and tetracycline with rather low tolerance for cadmium and copper. These observations indicate that visceral organs provide better conditions for bacterial growth and biological activity than exposed organs such as gill. Increase in the MIC of toxic metals as well as antibiotic resistance among aquatic bacterial populations is also an indication of risk to the safety of the aquatic ecosystem, fish fauna, and ultimately human health.
MAR values were ranging from 0.2 to 0.7. MAR indexes of the present work revealed that bacteria from locally raised fish may has been exposed to test antibiotics. McPhearson et al., 1991 reported that the MAR index of bacteria from a catfish pond, near a river where antibiotic was commonly used as treatment, was as high as 0.76.
Currently, local fish farmers employ amoxicillin to treat fish diseases. The present results proven that amoxicillin is no longer effective, since maximum bacterial isolates were sensitive to it. It appears that the emergence of resistance is also influenced by the physicochemical characteristics of water and several environmental factors including hospital and aquaculture waste disposal (Rhodes et al., 2000; Iwu 2020) along with the form and bioavailability of metals in the ecosystem. Resistance also develops from a non-specific mechanism with gene regulation of plasmids and chromosomes which may be heritable due to the presence of a resistance factor (R-factor) among the aquatic bacterial population (Silver and Walderhaug, 1992). The infections caused by the pathogenic bacteria with R-plasmids may pose a risk of therapeutic problems to public health and fish population. Thus, the water bodies with antibiotic and metal resistant bacteria serve as an environmental reservoir and source for the development of this trait among opportunistic pathogens and constitute a significant public concern. Therefore, such studies should be considered for the selection of antibiotics in dealing with water-borne diseases, particularly among fishermen and fish consumers. These findings indicate that sewage and industrial pollution are responsible for the emergence of bacterial resistance and deterioration of water quality, along with risk to biodiversity of the hydrobionts and the human health.
Conflict of Interest: We declare that we have no conflict of interest
Ethical Approval: This article is not under consideration or published elsewhere. Ethical clearance for the study was obtained from IAEC, Approval No. 17/IAEC (05)/RNLKWC/2019, Dated 27.07.2019.
ACKNOWLEDGEMENTS
Authors are grateful to Principal, PG Department of Zoology, Raja Narendra Lal Khan Women’s College (Autonomous) Natural and Applied Science Research Centre, Paschim Medinipore, West Bengal, India and Teacher-In-Charge, Dr BNDS Mahavidyalaya, Hagobindapur for the facilities provided and acknowledge the support to AD to carry out the paper as a part of the thesis under Vidyasagar University, Raja Narendra Lal Khan Women’s College (Autonomous).
REFERENCES
Ahmad R, Pandey RB, Arif SH, Nabi N, Jabeen M, Hasnain A (2012). Plymorphic β and γ lens crystallines demonstrate latitudinal distribution of threatened walking cat fish Clarias batrachus (Linn.) populations in North-western. Indian J Biol. Sci ; 12: 98-104.
Bauer AW, Kirby WM, Sherris JC, Turck M (1966). Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol; 45(4):493-496.
Binoy VV (2010). Catfish Clarias is vanishing from the waters of Kerala. Curr Sci; 99: 714.
Boateng TK, Opoku F, Akoto O (2019). Heavy metal contamination assessment of ground water quality: a case study of Oti landfill site, Kumasi. Applied Water Science; 9:33. 10.1007/s13201-019-0915-y.
Chapman JS (2003). Disinfectant resistance mechanisms, cross resistance, and co-resistance. Int Biodeter Biodegr; 51: 271–276.
Chatterjee SK, Bhattacharjee I, Chandra G (2010). Water quality assessment near an industrial site of Damodar River, India. Environ Monit Assess; 161: 177-189, DOI 10.1007/s10661-008-0736-1.
Clinical and Laboratory Standards Institute (2017). Performance Standards for Antimicrobial Susceptibility Testing (27th ed.). Wayne, PA, USA:
Edition Informational Supplement M100-S27. CLSI.
Cowan ST, Steel KJ (1993). Manual for the Identification of Medical Bacteria, Cambridge University Press, Cambridge.
Debnath S (2011). Clarias batrachus, the medicinal fish: An excellent candidate for aquaculture and employment generation. Proceedings of International Conference on Asia agriculture an animal, July 2-3, 2011, Hongkong.
Fattal B, Dotan A, Parpari L, Techorsh Y, Cabelli VJ (1993). Microbiological purification of fish grown in fecally contaminated commercial fish pond. Water Sci Technol; 27: 303-311.
Frost L, Leplae R, Summers A, Toussaint A (2005). Mobile genetic elements: The agents of open source evolution. Nature Rev Microbiol; 3: 722– 732.
Genchi G, Sinicropi MS, Lauria G, Carocci A, Catalano A (2020). The effects of Cadmium toxicity. Int J Environ Res Public Health ; 17: 3782; doi:10.3390/ijerph17113782.
Goswami B (2007). Magur (Clarias batrachus) seed production using low hatcheries. Aquacult Asia Mag; 12: 14-16.
Hossain Q, Hossain MA, Parween S (2006.) Artificial breeding and nursery practices of Clarias batrachus (Linnaeus, 1758). Sci World; 4:32-37.
Iloms E, Ololade OO, Ogola HJO, Selvarajan R (2020). Investigating Industrial Effluent Impact on Municipal Wastewater Treatment Plant in Vaal, South Africa. Int J Environ Res Public Health; 17: 1096; doi:10.3390/ijerph17031096.
Iwu CD, Korsten L, Okoh AL (2020). The incidence of antibiotic resistance within and beyound the agricultural ecosystem: A concern for Public Health. MicrobiologyOpen; .;9:e1035. doi.org/10.1002/mbo3.1035.
Kinare P, Shingadia H (2014). Screening of heavy metal resistant bacteria from Nale Lake of Vasai Taluka of Maharashtra. Int J Life Sci; 2(2):139–142.
Kobya M, Demirbas E, Senturk E, Ince M (2005). Adsorption of heavy metal ions from aqueous solutions by activated carbon prepared from apricot stone. Bioresour Technol; 96: 1518–1521.
Krumperman PH (1983). Multiple antibiotic resistance indexing of E. coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol; 46(1): 165-170.
Labrador KL, Nacario MAG, Malajacan GT, Abello JJM, Galarion LH, Rensing C, Rivera WL (2020). Selecting rep-PCR markers to source track fecal contamination in Laguna Lake, Philippines J Water Health; 18 (1): 19–29.
Liao XP, Tang W, Zhou RQ, Shi B (2008). Adsorption of metal anions of vanadium(V) and chromium(VI) on Zr(IV)-impregnated collagen fiber. Adsorption; 14: 55–64.
MacMillan JR (2001). Aquaculture and antibiotic resistance: a negligible public health risk?. World Aquacult; 32: 49-50.
Malik A, Ahemad M (2006). Genotoxicity of some wastewaters in India. Environ Toxicol Water Qual; 10(4):287–293.
Markey B, Leonard F, Archambault M, Cullinane A, Maguire D (2013). Clinical Veterinary Microbiology, Oxford University Press, New York, NY, USA, 2nd edition.
McMillan EA, Gupta SK, Williams LE, Jove T, Hiott LM, Woodley TA, Barrett JB, Jackson CR, Wasilenko JL, Simmons M, Tillman GE, McClelland M, Frye JG (2019). Antimicrobial Resistance Genes, Cassettes, and Plasmids Present in Salmonella enterica Associated With United States Food Animals. Front Microbiol; doi.org/10.3389/fmicb.2019.00832.
McPhearson RM, DePaola A, Zywno SR, Motes ML, Guarino AM (1991). Antibiotic resistance in gram-negative bacteria from cultured catfish and aquaculture ponds. Aquaculture; 99: 203-211.
Pathak SP, Bhattacharjee JW, Ray PK (1993). Seasonal variation in survival and antibiotic resistance among various bacterial populations in a tropical river. J Gen Appl Microbiol; 39: 47-56.
Pathak SP, Gopal K (2005). Occurrence of antibiotic and metal resistance in bacteria from organs of river fish. Environ. Res; 98: 100–103.
Priyadarshini SL, Suresh M, Huisingh D (2020). What can we learn from previous pandemics to reduce the frequency of emerging infectious diseases like COVID-19? Global Transitions; 2:202-220.
Rhodes G, Huys G, Swings, J, McGann P, Hiney M, Pickup RW (2000). Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: Implication of xTn 1721 in dissemination of the tetracycline resistance determinant Tet A. Appl. Environ. Microbiol; 66: 3883-3890.
Roy D, Mitra A, Biswas M, Chakraborty S, Pal S, Homechaudhuri S (2019). Early ontogeny of the Asian catfish Magur, Clarias batrachus (Linnaeus, 1758). Int J Fish Aquatic Stud; 7(1): 287-292.
Silver S, Walderhaug M (1992). Gene regulation of plasmid and chromosomes determined inorganic ion transport in bacteria. Microbiol. Rev; 5: 195-228.
Staley JT, Bryant MP, Pfenning N, Holt JG (1989). Bergey’s Manual of Systematic
Bacteriology, vol. 3, Williams and Wilkins Co., Baltimore,pp. 1582–
2298.
Tham LG, Perumal N, Syed MA, Shamaan MA, Shukor MY (2009). Assessment of Clarias batrachus as a source of acetylcholineesterase (AchE) for the detection of insecticides. J Environ Biol; 30: 135-138.
Venner S, Feschotte C, Biemont C (2009). Dynamics of transposable elements: Towards a community ecology of the genome. TIG: 25: 317–323.
Wamala SP, Mugimba KK, Mutoloki S, Evensen Ø, Mdegela R, Byarugaba DK, Sørum H (2018). Occurrence and antibiotic susceptibility of fish bacteria isolated from Oreochromis niloticus (Nile tilapia) and Clarias gariepinus (African catfish) in Uganda. Fisheries and Aquatic Sciences; 21:6, doi 10.1186/s41240-017-0080-x.
Wang XJ, Tao S (1998). Spatial structures and relations of heavy metal content in wastewater irrigated agricultural soil of Beijing’s Eastern farming regions. B. Environ Contam Tox; 61: 261–268.
Wiggins BA, Andrews RW, Conway RA, Dobratz CL, Dougherty DP, Eppard JR, Knupp SR, Limjoco MC, Sonsino J, Torrijos R, Zimmerman ME (1999). Use of antibiotic resistance analysis to identify nonpoint sources of faecal pollution. Appl Environ Microbiol; 65: 3427-3432.