1Department of Biotechnology, UIET, Maharshi Dayanand University Rohtak, Haryana 2School of Life Sciences, IIMT University, Meerut, India.
Corresponding author email: lko456@rediffmail.com
Article Publishing History
Received: 12/12/2020
Accepted After Revision: 23/03/2021
Pyruvate kinase M2 isoform (PKM2) in a less active state (dimer form) regulates the rate-limiting step of glycolysis that switches the glucose metabolism to aerobic glycolysis in tumor cells and thus promotes cell proliferation. Allosteric regulated PKM2 switches low to high activity state and prevent growth of cancer. Activators of PKM2 promote tetramer formation and suppress tumorigenesis. We present a structure based virtual screening of a diverse chemical compound collection (Diverse-lib) to identify novel activators of Pyruvate kinase M2 (PDB ID: 4G1N) from Homo sapiens. In order to rank potential small molecule hits, two separate docking algorithms were used to produce a consensus score. Four compounds out of 99,288 leads having lowest binding affinity even lower than control NZT compound were identified as activators of PKM2 using MTiOpenScreen and MTiAutoDock servers.
Further, these best predicted compounds were subjected for physicochemical, pharmacokinetic and toxicological investigation using preADMET tool and cross verified by SwissADME tool. Compound PubChem SID 17517397was satisfied all the ADME/Tox parameters out of four activators. In the AutoDock Vina and AutoDock programmes, the binding energy of compound SID 17517397 was -10 kcal / mol and -11.11 kcal/mol with PKM2.Compound SID 17517397 had human intestinal absorption, Caco2 cell permeability, Plasma Protein Binding and Blood-Brain Barrier penetration values as 94.18%, 23.86, 89.50%, 3.30, respectively, which indicates that it is in the range of well absorbed and active compound range. Compound had negative carcinogenicity value in mouse and rat. Therefore, it is concluded that compound could be promising novel activator for PKM2 as drug target but it must be verified by experimental studies.
ADMET/Tox, Cancer, Mtiautodock, Mtiopenscreen, Pyruvate Kinase M2,
Sharma P, Singh M, Sharma S. Analysis on In-silico Identification of Novel Activator of Pyruvate Kinase M2 . Biosc.Biotech.Res.Comm. 2021;14(1).
Sharma P, Singh M, Sharma S. Analysis on In-silico Identification of Novel Activator of Pyruvate Kinase M2 . Biosc.Biotech.Res.Comm. 2021;14(1). Available from: <a href=”https://bit.ly/397OnBq“>https://bit.ly/397OnBq</a>
Copyright © Sharma et al., This is an open access article distributed under the terms of the Creative Commons Attribution License (CC-BY) https://creativecommns.org/licenses/by/4.0/, which permits unrestricted use distribution and reproduction in any medium, provide the original author and source are credited.
INTRODUCTION
In cancer cells, the metabolism differs significantly from that of healthy cells (Kim and Dang, 2006; Vander Heiden et al., 2009; Zhao et al., 2013). In normal cell rely on glycolysis to produce energy but in tumor cells it switches the glucose metabolism to aerobic glycolysis and this mechanism is called Warburg effect (Warburg, 1956; DeBerardinis et al., 2008). Tumor’s glycolysis interventions are a novel approach for targeted anti-cancer therapies (Chen et al., 2007; Gatenby and Gillies, 2007; Porporato et al., 2011).
In cancer cell metabolism, the regulation of Pyruvate kinase M2 isoform (PKM2) plays a key role. The last rate-limiting enzyme in the glycolytic pathway is pyruvate kinase (PK), which catalyses the transfer of a phosphate group from phosphoenolpyruvate to ADP to obtain pyruvate and ATP. There are four distinct subtypes of Pyruvate kinase. PKL isoforms exist predominantly in the liver, kidney and red blood cells, whereas PKR is mostly present in red blood cells (Gupta and Bamezai, 2010; Israelsen et al., 2013, Wong et al., 2013; Yang and Lu, 2013; Otto et al., 2016).
In myocardium, skeletal muscle and brain tissue, PKM1 is distributed, and in tissues such as the brain and liver, PKM2 is distributed (Israelsen et al., 2013). For cancer metabolism and tumour development, PKM2 is essential, yet tetramer and dimer of PKM2 consist of the same monomer (Ashizaw et al., 1991; Yang and Lu, 2015). There are substantially different biological effects between the tetramer and dimer form (Muñoz-Colmenero et al., 2015).
In the sense of glucose metabolism, the tetramer primarily plays the role of pyruvate kinase and controls glycolysis and dimer PKM2 as a switch for energy metabolism and material synthesis (Dombrauckas et al., 2005). In the dimer state, PKM2 can enter the nuclear to regulate gene expression, epithelial–mesenchymal transition (EMT), invasion and metastasis and cell proliferation. Zhang et al. (2019) first used the switching effect of PKM2 in glucose metabolism to expand and enrich the Warburg effect (Zhang et al., 2019).
Endogenous and exogenous activators allosterically regulate the change between PKM2 dimers and tetramers. PKM2 dimers are tetramerized using activators that allow PKM2 to behave like PKM1 induces reversal of the Warburg effect in cancer cells. Phosphorylation or acetylation of native tetrameric PKM2 in cancer cells causes a transition to a dimeric/monomeric form, which translocate into the nucleus and causes oncogene transcription. However, it is unclear how these post-translational modifications (PTMs) cause PKM2 to lose its oligomeric state. Nandi et al.
(2020) performed crystallographic and biophysical studies of PKM2 mutants containing residues that mimic phosphorylation and acetylation Nandi et al. (2020). They discovered that PTMs cause a significant structural reorganization of the binding site for fructose 1, 6-bisphosphate (FBP), an allosteric activator, affecting the interaction with FBP and causing oligomerization disruption (Nandi et al., 2020).
In the current research, we have implemented a virtual screening approach to search out the novel activator for the treatment of cancer targeting the PKM2 protein. In-silico physicochemical, pharmacokinetic and toxicological properties of activators were analysed by preADMET and cross-checked with the SwissADME web tool (Lee et al., 2003; Daina et al., 2017).
MATERIAL AND METHODS
The 3D crystal structure of Pyruvate kinase isoform M2 in complex with an activator was retrieved from the Protein Data Bank with PDB ID: 4G1N (Kung et al., 2012). All the water molecules, Oxalate, Magnesium ions and N-(4-{[4-(pyrazin-2-yl) piperazin-1-yl] carbonyl} phenyl) quinoline-8-sulfonamide (NZT) were removed and polar hydrogen added to PKM2 protein for structure based virtual screening as well as molecular docking processes. Small-molecule activator NZT bind PKM2 at the subunit interaction interface, a site that is distinct from fructose-1,6-bisphosphate (FBP) was used as activator binding site in molecular docking (Kung et al., 2012).
The virtual screening was performed using the diverse chemical compound library (Diverse-lib) database. The compound library consisted of 99,288 diverse drug-like PubChem compounds from in house Diverse-lib database of RPBS Web portal. The compound library was filtered by using the criteria such as molecular weight <500 Dalton; hydrogen bond donor <5, hydrogen bond acceptor <10, octanol–water partition coefficient logP <5; Number of rotatable bonds <8, polar surface area <140 Å (Labbé et al., 2015).
Docking was carried out on the MTiOpenScreen server with AutoDock Vina and with AutoDock on MTiAutoDock server (Labbé et al., 2015). The rankings of AutoDock Vina and AutoDock were combined to construct a consensus list of compounds with both techniques that scored well. Validation and Optimization process of best predicted compounds had been processed by PreADMET tool and cross check by Swiss ADME tool, which are web-based application for predicting absorption, distribution; metabolism, elimination and toxicity (Lee et al., 2003; Daina et al., 2017). Using Python Molecular Viewer software, docking findings were visualized to display the 3D structure and position of activator binding to the protein (Sanner, 1999; Daina et al., 2017).
RESULTS AND DISCUSSION
Activator binding site analysis: Small molecule activator PubChem SID 17517397 bind PKM2 at the subunit interaction interface, a site different from that of the endogenous activator fructose-1, 6-bisphosphate (FBP). The activator binding sites of PKM2 comprises 8 residues on chain A such as TYR390, ASP354, GLN393, ILE389, PHE26, GLU397, LEU353, LEU394 and 7 residues on chain B as MET30, PHE26, TYR390, LYS311, LEU394, LEU353, ASP354. Binding of activator to PKM2 promoted a constitutively active enzyme state (tetramer form).
Structure based virtual screening: 1500 small molecules were screened after applying filter criteria in database. A gradient-based conformational search approach was employed by AutoDock Vina. The grid box parameters were set to values of 3.743 Å,-12.72, and 48.977 Å for the grid box center and 34 Å ×28 Å ×32 Å for the box dimensions. We use a total of 10 binding modes and 8 for exhaustiveness. The scoring of the docking poses produced and the ranking of the ligands were based on the empirical scoring function of Vina approximating the affinity of binding in kcal/mol. Top 100 compounds were selected based on their lowest binding energy and further docked with activator binding site of PKM2 using AutoDock. Docking results of both were shown in table 1.
Table 1. Docking results of Autodock Vina and Autodock programmes with physicochemical properties of compounds.
| Sl. No | PubChem SID | BE (AutoDock Vina) | BE (AutoDock) | nRot | HBA | HBD | LogP | MW | TPSA |
| 1 | 17517397 | -10 | -11.11 | 6 | 4 | 2 | 3.98 | 374.9043 | 58.2 |
| 2 | 26649876 | -9.9 | -11.34 | 6 | 6 | 1 | 3.29 | 374.4357 | 72.95 |
| 3 | 24838666 | -9.6 | -9.25 | 4 | 7 | 2 | 3.11 | 400.8587 | 99.94 |
| 4 | 49737693 | -9.5 | -11.28 | 8 | 7 | 1 | 3.65 | 413.8542 | 86.48 |
| 5 | 24339460 | -9.5 | -7.35 | 2 | 7 | 2 | 0.18 | 193.1628 | 96.51 |
| 6 | 4247715 | -9.5 | -11.71 | 6 | 7 | 1 | 3.77 | 398.4108 | 118.38 |
| 7 | 24798741 | -9.4 | -9.38 | 7 | 7 | 0 | 1.98 | 340.3764 | 85.26 |
| 8 | 4256388 | -9.4 | -9.34 | 1 | 4 | 0 | 3.1 | 218.2981 | 43.6 |
| 9 | 17477384 | -9.4 | -9.47 | 4 | 5 | 3 | 2.95 | 352.4069 | 97.64 |
| 10 | 26664864 | -9.2 | -8.57 | 1 | 5 | 0 | 1.63 | 204.1821 | 57.38 |
| 11 | 49736489 | -9.2 | -8.92 | 2 | 4 | 0 | 3.98 | 338.7876 | 46.34 |
| 12 | 17469113 | -9.2 | -8.24 | 3 | 4 | 1 | 2.75 | 260.1661 | 52.6 |
| 13 | 24779480 | -9.2 | -10.05 | 6 | 7 | 2 | 2.68 | 360.4108 | 153.7 |
| 14 | 17479262 | -9.1 | -9.95 | 5 | 6 | 0 | 3.88 | 295.2927 | 84.74 |
| 15 | 56317084 | -9.1 | -9.31 | 4 | 6 | 0 | 2.22 | 256.2136 | 78.36 |
| 16 | 26534815 | -9.1 | -9.85 | 2 | 5 | 0 | 3.39 | 317.386 | 103.68 |
| 17 | 7971621 | -9.1 | -8.93 | 3 | 7 | 1 | 1.52 | 331.3464 | 107.75 |
| 18 | 22413776 | -9 | -9.75 | 2 | 5 | 0 | 3.62 | 253.256 | 63.12 |
| 19 | 49716534 | -8.9 | -9.62 | 6 | 7 | 1 | 2.23 | 392.4477 | 113.63 |
| 20 | 49817894 | -8.9 | -8.26 | 6 | 6 | 1 | 2.41 | 356.2965 | 73.59 |
| 21 | 26537356 | -8.9 | -8.70 | 3 | 4 | 2 | 3.97 | 337.8028 | 57.78 |
| 22 | 17449251 | -8.9 | -10.08 | 6 | 7 | 1 | 3.88 | 369.3944 | 128.51 |
| 23 | 89852923 | -8.9 | -8.00 | 2 | 6 | 1 | 0.97 | 190.1588 | 87.39 |
| 24 | 862504 | -8.9 | -8.81 | 4 | 6 | 0 | 3.29 | 330.3401 | 73.93 |
| 25 | 849520 | -8.9 | -10.84 | 8 | 7 | 2 | 2.36 | 386.4216 | 106.02 |
| 26 | 4242876 | -8.9 | -8.57 | 2 | 3 | 1 | 3.49 | 275.3013 | 42.24 |
| 27 | 22402150 | -8.8 | -9.88 | 4 | 7 | 1 | 3.01 | 352.3672 | 121.87 |
| 28 | 26620089 | -8.8 | -11.71 | 6 | 6 | 1 | 3.18 | 414.5178 | 84.09 |
| 29 | 7967102 | -8.8 | -9.46 | 4 | 6 | 0 | 3.44 | 349.1363 | 78.36 |
| 30 | 57263884 | -8.8 | -10.18 | 10 | 7 | 2 | 2.99 | 372.415 | 97.64 |
| 31 | 24784913 | -8.8 | -9.11 | 6 | 6 | 0 | 3.24 | 376.4516 | 62.47 |
| 32 | 26650965 | -8.8 | -11.12 | 5 | 7 | 0 | 3.04 | 394.399 | 80.82 |
| 33 | 49674809 | -8.8 | -6.59 | 7 | 6 | 2 | 3.5 | 376.4052 | 95.15 |
| 34 | 24296145 | -8.8 | -8.14 | 2 | 7 | 1 | 3.1 | 323.3293 | 123.89 |
| 35 | 26725965 | -8.8 | -10.68 | 5 | 7 | 2 | 3.51 | 420.3899 | 102.68 |
| 36 | 17447282 | -8.8 | -8.45 | 2 | 4 | 1 | 3.59 | 305.7362 | 84.61 |
| 37 | 24831749 | -8.7 | -9.88 | 6 | 5 | 0 | 3.53 | 335.3533 | 65.49 |
| 38 | 26649874 | -8.7 | -9.13 | 6 | 6 | 2 | 3.03 | 368.4263 | 76.66 |
| 39 | 7964748 | -8.7 | -10.28 | 5 | 7 | 1 | 3.03 | 375.442 | 111.78 |
| 40 | 17456133 | -8.7 | -8.24 | 4 | 6 | 3 | 2.55 | 329.3736 | 103.01 |
| 41 | 26647975 | -8.7 | -8.81 | 3 | 5 | 0 | 3.79 | 378.4641 | 43.21 |
| 42 | 26648535 | -8.7 | -7.27 | 5 | 5 | 1 | 3.47 | 353.4116 | 56.79 |
| 43 | 24809269 | -8.7 | -8.75 | 6 | 6 | 1 | 3.45 | 300.3092 | 84.15 |
| 44 | 17465009 | -8.7 | -6.39 | 1 | 4 | 2 | 3.69 | 329.3751 | 104.53 |
| 45 | 26533701 | -8.7 | -7.74 | 2 | 4 | 0 | 2.64 | 280.2021 | 48.03 |
| 46 | 26727266 | -8.7 | -9.89 | 4 | 6 | 0 | 3.29 | 327.3313 | 81.35 |
| 47 | 14730768 | -8.7 | -9.50 | 5 | 5 | 0 | 3.3 | 336.338 | 73.58 |
| 48 | 49737701 | -8.7 | -9.69 | 6 | 7 | 1 | 1.75 | 346.3777 | 82.82 |
| 49 | 24364860 | -8.7 | -6.77 | 3 | 4 | 2 | 2.98 | 296.3636 | 66.13 |
| 50 | 93577670 | -8.7 | -10.07 | 7 | 7 | 1 | 2.03 | 351.356 | 94.31 |
| 51 | 49645683 | -8.6 | -8.94 | 4 | 6 | 1 | 2.96 | 296.2775 | 88.28 |
| 52 | 17470642 | -8.6 | -9.50 | 5 | 5 | 2 | 3.37 | 361.3906 | 67.79 |
| 53 | 4246955 | -8.6 | -8.87 | 4 | 7 | 2 | 3.63 | 338.4069 | 84.73 |
| 54 | 17512468 | -8.6 | -7.58 | 1 | 5 | 4 | 3.06 | 284.3363 | 112.04 |
| 55 | 14744922 | -8.6 | -9.57 | 7 | 6 | 1 | 2.89 | 387.4494 | 97.92 |
| 56 | 26648015 | -8.6 | -7.70 | 7 | 7 | 2 | 2.02 | 390.4814 | 88 |
| 57 | 860027 | -8.6 | -7.02 | 2 | 4 | 0 | 3.48 | 362.4448 | 67.2 |
| 58 | 11534585 | -8.6 | -9.51 | 3 | 5 | 0 | 3.1 | 261.2813 | 67.39 |
| 59 | 851712 | -8.6 | -9.26 | 4 | 6 | 0 | 1.56 | 274.2984 | 103.14 |
| 60 | 16952534 | -8.6 | -8.96 | 4 | 5 | 1 | 3.96 | 297.3053 | 72.56 |
| 61 | 17402129 | -8.5 | -7.61 | 2 | 4 | 1 | 3.43 | 319.3324 | 58.11 |
| 62 | 24811377 | -8.5 | -11.15 | 7 | 7 | 0 | 3.64 | 350.3249 | 98.15 |
| 63 | 124949736 | -8.5 | -8.04 | 6 | 5 | 2 | 3.57 | 406.3982 | 67.43 |
| 64 | 24334833 | -8.5 | -9.20 | 3 | 5 | 1 | 3.97 | 292.2888 | 78.94 |
| 65 | 24293703 | -8.5 | -10.63 | 3 | 7 | 1 | 3.02 | 289.2667 | 129.19 |
| 66 | 24366734 | -8.5 | -10.40 | 4 | 4 | 1 | 3.98 | 327.7617 | 59.31 |
| 67 | 49640535 | -8.5 | -7.27 | 6 | 7 | 2 | 2.78 | 384.3793 | 97.28 |
| 68 | 57259063 | -8.5 | -7.65 | 5 | 6 | 1 | 2.76 | 327.3313 | 73.86 |
| 69 | 24807319 | -8.5 | -6.83 | 5 | 7 | 1 | 1.42 | 300.2662 | 86.75 |
| 70 | 24412722 | -8.5 | -5.59 | 7 | 7 | 1 | 2.08 | 434.5108 | 119.25 |
| 71 | 26729749 | -8.5 | -7.86 | 3 | 5 | 1 | 1.68 | 210.6204 | 63.5 |
| 72 | 144203702 | -8.5 | -5.71 | 9 | 5 | 2 | 3.85 | 414.3427 | 64.17 |
| 73 | 4247822 | -8.5 | -7.67 | 4 | 6 | 2 | 3.42 | 400.473 | 71.33 |
| 74 | 49718523 | -8.5 | -8.99 | 5 | 6 | 0 | 3.47 | 383.484 | 60.01 |
| 75 | 89855761 | -8.5 | -7.13 | 1 | 5 | 1 | 1.08 | 195.2184 | 68.01 |
| 76 | 3714881 | -8.5 | -10.69 | 6 | 6 | 0 | 3.84 | 365.4057 | 99.48 |
| 77 | 14720958 | -8.5 | -9.47 | 8 | 5 | 0 | 3.82 | 404.5014 | 57.69 |
| 78 | 17445644 | -8.5 | -9.34 | 5 | 5 | 1 | 3.9 | 336.2653 | 74.92 |
| 79 | 26652683 | -8.4 | -8.94 | 3 | 4 | 1 | 3.13 | 336.3612 | 93.48 |
| 80 | 24787424 | -8.4 | -9.58 | 4 | 7 | 1 | 1.19 | 234.2114 | 92.96 |
| 81 | 17452005 | -8.4 | -8.47 | 5 | 7 | 2 | 3.42 | 359.3813 | 96.02 |
| 82 | 4247583 | -8.4 | -9.82 | 6 | 7 | 2 | 1.22 | 388.4639 | 137.67 |
| 83 | 7977979 | -8.4 | -10.98 | 8 | 6 | 1 | 3.76 | 383.464 | 102.55 |
| 84 | 49733540 | -8.4 | -10.37 | 5 | 6 | 2 | 2.97 | 385.3905 | 108.86 |
| 85 | 24834152 | -8.4 | -8.08 | 6 | 6 | 3 | 2.43 | 387.4958 | 100.88 |
| 86 | 24348650 | -8.4 | -10.17 | 6 | 7 | 1 | 3.94 | 383.7852 | 97.04 |
| 87 | 24801340 | -8.4 | -7.24 | 2 | 4 | 0 | 2.74 | 277.2741 | 52.21 |
| 88 | 24365159 | -8.4 | -9.17 | 3 | 5 | 0 | 2.3 | 246.2619 | 67.41 |
| 89 | 847046 | -8.4 | -8.00 | 5 | 6 | 0 | 2.26 | 252.6571 | 69.9 |
| 90 | 104223077 | -8.4 | -9.72 | 8 | 6 | 2 | 1.78 | 354.3997 | 88.4 |
| 91 | 14722665 | -8.4 | -9.49 | 4 | 7 | 1 | 1.67 | 280.2566 | 122.81 |
| 92 | 17506536 | -8.4 | -9.37 | 4 | 6 | 0 | 2.82 | 292.3303 | 75.36 |
| 93 | 24824418 | -8.4 | -10.57 | 1 | 6 | 2 | 1.69 | 293.2768 | 84.08 |
| 94 | 24320463 | -8.4 | -9.80 | 4 | 7 | 1 | 2.78 | 357.2846 | 98.39 |
| 95 | 24798468 | -8.4 | -10.25 | 6 | 6 | 2 | 2.65 | 338.4036 | 83.81 |
| 96 | 85269299 | -8.4 | -10.17 | 8 | 7 | 1 | 3 | 332.3511 | 101.22 |
| 97 | 26618413 | -8.4 | -8.21 | 4 | 7 | 2 | 1.03 | 383.4673 | 112.7 |
| 98 | 17515744 | -8.4 | -11.00 | 5 | 6 | 1 | 3.55 | 339.3684 | 116.05 |
| 99 | 24782536 | -8.4 | -10.26 | 3 | 7 | 2 | 1.94 | 272.2594 | 99.84 |
| 100 | 17453955 | -8.3 | -9.17 | 1 | 6 | 2 | 1.1 | 242.2334 | 83.54 |
BE: binding energy (kcal/mol), nRot:number of rotatable bonds, HBA: hydrogen bond donor Count, HBD: hydrogen bond acceptor Count, LogP: partition coefficient of a molecule between an aqueous and lipophilic phase, MW: molecular weight, TPSA:topological polar surface area.
Four compounds with PubMed SID 17517397, 26649876, 49737693, 4247715 were selected based on binding energy of compound with PKM2 protein having >-9.5kcal/mol in AutoDock Vina and >-11kcal/mol in AutoDock, which is even lower than control NZT compound (binding energy: -7.95kcal/mol with PKM2 protein).
ADME/Tox properties of best predicted compounds: The properties of human intestinal absorption were critical for the production of drugs that purport to be orally administered (Zhao et al., 2001; Postigo et al., 2010). Human intestinal absorption (%HIA) values of four best predicted compounds with PubMed SID 17517397, 26649876, 49737693 and 4247715 were shown in table 2. These compounds have been identified in the category of well absorbed compounds (HIA: 70 ~ 100 %) (Yee, 1997). In vitro cell permeability Caco-2 is an important test that measures drug intestinal absorption (Yazdanian et al., 1998).
In the MDCK method, the cell permeability in vitro used canine kidney cells and has a shorter growth rate than the Caco-2 cells were used as a tool for the rapid analysis of permeability (Irvine et al., 1999). In the pharmaceutical industry, skin permeability is used to measure the toxicity of chemical products in the event of unintended skin touch (Singh and Singh, 1993). Moreover, the blood-brain barrier (BBB) was essential for drug pharmacology. PCaco-2, MDCK, Skin Permeability, PPB and BBB of four compounds were shown in table 2. Compound SID 26649876 had a BBB value greater than 2.0 and was graded in the central nervous system with high absorption (Ma et al., 2005).
Table 2. ADMET properties of best predicted activators.
| Pubchem SID | PCaco-2 (nm/sec) | MDCK
(nm/sec) |
Skin Permeability | %HIA | %PPB | BBB | Carcinogenicity | ||
| Ames test | Mouse | Rat | |||||||
| 17517397 | 23.86 | 0.11 | -3.91 | 94.18 | 89.50 | 3.30 | mutagen | -ve | -ve |
| 26649876 | 31.32 | 0.13 | -3.25 | 96.35 | 99.32 | 0.19 | mutagen | -ve | +ve |
| 49737693 | 26.33 | 1.55 | -3.80 | 96.77 | 87.55 | 0.02 | mutagen | -ve | -ve |
| 4247715 | 19.85 | 0.84 | -4.13 | 97.98 | 92.05 | 0.18 | mutagen | -ve | -ve |
PPB:Plasma Protein Binding, BBB: blood brain barrier.
Carcinogenicity is the toxicity in the body that causes cancer. Compounds SID 26649876 had positive carcinogenicity value in rat. Compounds SID 26649876, 49737693 and 4247715 had a BBB value less than 2.0 and was graded in the central nervous system with low absorption (Ma e al., 2005). Further, ADME properties of compounds PubChem SID17517397, 26649876, 49737693 and 4247715 were analysed by SwissADME tool (Daina et al., 2017). All the compounds were qualified five different rule-based filters such as Lipinski filter implemented rule-of-five, Ghose, Veber, Egan and Muegge rules shown in table 3 (Ghose et al., 1999; Egan et al., 2000; Muegge et al., 2001; Veber et al., 2002; Lipinski, 2004). The result provided in table 3 shows that all the compounds investigated have high gastrointestinal absorption and good skin permeation.
Table 3. The pharmacokinetics and drug likeliness prediction of four best predicted compounds
| Pubchem SID | GI | BBB | Log Kp (skin permeation Coefficient) (cm/s) | Bioavalia-bility Score | Lipinski | Ghose | Veber | Egan | Muegge |
| 17517397 | High | Yes | -5.76 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 26649876 | High | Yes | -6.25
|
0.55 | Yes | Yes | Yes | Yes | Yes |
| 49737693 | High | No | -6.23 | 0.55 | Yes | Yes | Yes | Yes | Yes |
| 4247715 | High | No | -6.05 | 0.55 | Yes | Yes | Yes | Yes | Yes |
GI: Gastrointestinal absorption, BBB: Blood Brain Barrier penetration.
Compounds SID 49737693 and 4247715 indicate negative results in blood-brain barrier (BBB) permeation. The SwissADME estimates for passive human gastrointestinal absorption (GI) and permeation to the blood-brain barrier (BBB) consists of the BOILED-Egg model reading (Butina et al., 2002). Therefore, only one compound PubChem SID 17517397 qualifies all parameters of ADME /Tox on analysis of four best predicted compounds using PreADMET and SwissADME tools (Butina et al., 2002).
Li et al. (2018) used structure-based virtual screening to find successful activators targeting PKM2 in the ZINC database. ZINC08383544, a screened compound, promotes the development of PKM2 tetramers, effectively blocks PKM2 nuclear translocation, and inhibits tumour growth, suggesting that it may be a novel PKM2 activator (Li et al., 2018).
Visualization of protein-ligand interaction: Best docked complex was analyzed through Python Molecular Viewer for their interaction study shown in figure 1.
Figure 1: Docking pose of compound SID 17517397 on PKM2 protein structure. One H-bond was formed between amino acid LYS311 of protein Chain B with compound. Chain A and B binding residues of PKM2 protein were colored with pink and royal blue, respectively.
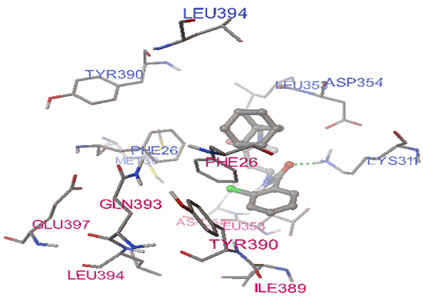
It is evident from this analysis that compound SID 1751739 was located at the subunit interaction interface of protein and was stabilized by hydrogen bonding.
CONCLUSION
The current research utilizes structure based virtual screening to identify human Pyruvate kinase M2 isoform (PKM2) protein activator that is required for cancer treatment. From thousands of chemical structures and a sequence of steps of rational refinement, including virtual screening, molecular docking and ADME/Tox studies, we identified compound PubChem SID 17517397 as novel activator of pyruvate kinase M2 protein as drug target for further experimental testing.
ACKNOWLEDGEMENTS
We would like to thank Professor Rahul Rishi, Director UIET, MDU, Rohtak, India and Dr. Sonia, HOD & Associate Professor, Department of Biotechnology, MDU, Rohtak, India for helping in preparation of the research paper.
Conflict of Interests: Authors declare no conflict of interests among themselves.
REFERENCES
Ashizaw, K., Willinghan, M.C., and Liang, C.M. (1991). In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem., 266, 16842-6.
Butina, D., Segall, M.D., and Frankcombe, K. (2002). Predicting ADME properties in silico: Methods and models. Drug Discov.Today. 7, S83–S88.
Chen, Z., Lu, W., Garcia-Prieto, C., and Huang, P. (2007). The Warburg effect and its cancer therapeutic implications. J Bioenerg Biomembr., 39, 267-74.
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep., 7, Article number: 42717.
DeBerardinis, R.J., Lum, J.J., Hatzivassiliou, G., and Thompson, C.B. (2008). The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11-20.
Dombrauckas, J.D., Santarsiero, B.D., and Mesecar, A.D. (2005). Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry, 44, 9417–29.
Egan, W. J., Merz, K.M., and Baldwin, J.J. (2000). Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 43, 3867–3877.
Gatenby, R.A., and Gillies, R.J. (2007). Glycolysis in cancer: a potential target for therapy. Int J Biochem Cell Biol., 39, 1358-66.
Ghose, A.K., Viswanadhan, V.N., and Wendoloski, J.J. (1999). A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1(1), 55–68.
Gupta, V., and Bamezai, R.N.K. (2010). Human pyruvate kinase M2: a multifunctional protein. Protein Sci., 19, 2031–44.
Irvine, J.D., Takahashi, L., Lockhart, K., Cheong, J., and Tolan, J.W. (1999). MDCK (Madin-Darby Canine Kidney) Cells: A Tool for Membrane Permeability Screening. Journal of Pharmaceutical Sciences. 88,28-33.
Israelsen, W.J., Dayton, T.L., Davidson, S.M., Fiske, B.P., Hosios, A.M., Bellinger, G., Li, J., Yu, Y., Sasaki, M., Horner, J.W., Burga, L.N., Xie, J., Jurczak, M.J., DePinho, R.A., Clish, C.B., Jacks, T., Kibbey, R.G., Wulf, G.M., Di Vizio, D., Mills, G.B., Cantley, L.C., and Vander Heiden, M.G. (2013). PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell., 155,397–409.
Kim, J.W., and Dang, C.V. (2006). Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res., 66, 8927–30.
Kung, C., Hixon, J., Choe, S., Marks, K., Gross, S., Murphy, E., DeLaBarre, B., Cianchetta, G., Sethumadhavan, S., Wang, X., Yan, S., Gao, Y., Fang, C., Wei, W., Jiang, F., Wang, S., Qian, K., Saunders, J., Driggers, E., Woo, H.K., Kunii, K., Murray, S., Yang, H., Yen, K., Liu, W., Cantley, L.C., Vander Heiden, M.G., Su, S.M., Jin, S., Salituro, F.G., and Dang, L. (2012). Small Molecule Activation of PKM2 in Cancer Cells Induces Serine Auxotrophy. Chem Biol., 19(9), 1187-1198.
Labbe, C. M., Rey, J., Lagorce, D., Vavrusa, M., Becot, J., Sperandio, O., Villoutreix, B.O., Tufféry, P., and Miteva, M.A. (2015). MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Research, 43(W1), W448-W454.
Lee, S.K., Lee, I.H., Kim, H.J., Chang, G.S., Chung, J.E., and No. K.T. (2003). The PreADME Approach: Web-based program for rapid prediction of physico-chemical, drug absorption and drug-like properties, EuroQSAR 2002 Designing Drugs and Crop Protectants: processes, problems and solutions, Blackwell Publishing, Massachusetts, USA. Pg 418-420.
Li, Y., Bao, M., Yang, C., Chen, J., Zhou, S., Sun, R., Wu, C., Li, X., Bao, J. (2018). Computer-aided identification of a novel pyruvate kinase M2 activator –compound. Cell Proliferation, 51, e125 09.
Lipinski, C.A. (2004). Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 1(4), 337–341.
Ma, X., Chen, C., and Yang, J. (2005). Predictive Model of Blood-Brain Barrier Penetration of Organic Compounds. Acta Pharmacologica Sinica., 26, 500-512.
Muegge, I., Heald, S.L., and Brittelli, D. (2001). Simple selection criteria for drug-like chemical matter. J. Med. Chem. 44, 1841–1846.
Muñoz-Colmenero, A., Fernández-Suárez, A., Fatela-Cantillo, D., Ocaña-Pérez, E., Domínguez-Jiménez, J.L., and Díaz-Iglesias, J.M. (2015). Plasma tumor M2-pyruvate kinase levels in different cancer types. Anticancer Res., 35, 4271-6.
Nandi, S., Razzaghi, M., Srivastava, D., and Dey, M. (2020). Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation. J. Biol. Chem., 295(51), 17425–17440.
Otto, A.M. (2016). Warburg effect(s)-a biographical sketch of Otto Warburg and his impacts on tumor metabolism. Cancer Metab. 4, Article number: 5.
Porporato, P.E., Dhup, S., Dadhich, R.K., Copetti, T., Sonveaux, P. (2011). Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Front Pharmacol., 2,49.
Postigo, M.P., Guido, R.V., Oliva, G., Castilho, M.S., da R Pitta, I., de Albuquerque, J.F., and Andricopulo, A.D. (2010). Discovery of New Inhibitors of Schistosoma mansoni PNP by Pharmacophore-Based Virtual Screening. Journal of Chemical Information and Modeling, 50, 1693-1705.
Sanner, M.F. (1999). Python: A Programming Language for Software Integration and Development. J Mol Graph Model. 17(1), 57-61.
Singh, S., and Singh, J. (1993). Transdermal Drug Delivery by Passive Diffusion and Iontophoresis: A Review. Medicinal Research Reviews, 13, 569-621.
Vander Heiden, M.G., Cantley, L.C., and Thompson, C.B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science, 324, 1029–33.
Veber, D.F., Johnson, S.R., Cheng, H.Y., Smith, B.R., Ward, K.W., and Kopple, K.D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem., 45, 2615–2623.
Warburg, O. (1956). On respiratory impairment in cancer cells. Science, 124,269–70.
Wong, N., De Melo, J., and Tang, D. (2013). PKM2, a Central Point of Regulation in Cancer Metabolism. Int J Cell Biol., Vol. 2013, Article ID 242513.
Yang, W., and Lu, Z. (2013). Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett., 339, 153-8.
Yang, W., and Lu, Z. (2015). Pyruvate kinase M2 at a glance. J Cell Sci., 128, 1655–60.
Yazdanian, M., Glynn, S.L., Wright, J.L., and Hawi (1998). Correlating Partitioning and Caco-2 Cell Permeability of Structurally Diverse Small Molecular Weight Compounds. Pharmaceutical Research. 15, 1490-1494.
Yee, S. (1997). In vitro permeability across Caco-2 cells (colonic) can predict in vivo (small intestinal) absorption in man-fact or myth. Pharm Res., 14(6), 763-6.
Zhang, Z., Deng, X., Liu, Y., Liu, Y., Sun, L., and Chen F. (2019). PKM2, function and expression and regulation. Cell Biosci 9, 52.
Zhao, Y., Butler, E.B., and Tan, M. (2013). Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis., 4:e532.
Zhao, Y.H., Le, J., Abraham, M.H., Hersey, A., Eddershaw, P.J., Luscombe, C.N., Butina, D., Beck, G., Sherborne, B., Cooper, I., and Platts, J.A. (2001). Evaluation of Human Intestinal Absorption Data and Subsequent Derivation of a Quantitative Structure-Activity Relationship (QSAR) with the Abraham Descriptors. Journal of Pharmaceutical Sciences. 90, 749-784.


