
Medical
Communication
Biosci. Biotech. Res. Comm. 10(4): 722-727 (2017)
Isolation and identi cation of pharmaceutically active
lipase producing
Bacillus
spp. from mangrove sediments
against Methicillin Resistant
Staphylococcus aureus
isolated from wound of patients
M.I. Farheena, A. Chithira and A. Mohankumar
Division of Microbial Technology, PG and Research Department of Zoology,
Chikkanna Govt. Arts College, Tirupur, Tamilnadu, India
ABSTRACT
Ninety two clinical wound samples were collected, among, 50 positive Staphylococcus aureus were isolated. These isolates were
characterized morphologically and biochemically. Twenty three antibiotics were used to determine susceptibility patterns by disc
diffusion method. The multiple antibiotic resistances (MAR) index were calculated according to the MAR index formula ranged
from 0.21 to 0.78. All the 50 isolates of MRSA were showed MAR index in between these ranges. Among these MAR index the
pathogenic isolates (100%) were resistant of Penicillin, while trimethoprim showed resistance (86%), cephoxitin (80%), kanamy-
cin (78%), vancomycin and cepfodoxamine (72%), moxalactam and quinupristin (70%), cotrimoxazole (66%), methicillin (64%),
novobiocin (62%) and erythromycin (56%) and there was no resistance found to chloramphenicol, and rifampicin. More than 65%
resistance MRSA isolates were selected for plasmid isolation. Natural products are boundless source for important novel com-
pounds having antagonistic activity against pathogenic organisms. Marine environment covers almost 70% of the earth surfaces.
Organisms present in these environments are extremely rich sources of bioactive compounds. The ocean remains as an unexploited
source of many drugs and pharmacologically active substances. Microbial enzymes have many advantages over the animal and
plant enzyme, rstly; they are economical and can be produced on large scale within the limited space and time. Secondly, they
are capable of producing a wide variety of enzymes; they can grow a wide range of environmental condition. Enzymes have many
roles in the pharmaceutical and diagnostic industries. The bacteria Bacillus spp. was isolated from the mangrove sediment, for the
lipase production. These enzymes may inhibit the growth of MRSA. Recently with the advent of biotechnology, there has been a
growing interest and demand for enzymes with the novel properties.
KEY WORDS: PUS,
STAPHYLOCOCCUS AUREUS
, ANTIBIOTIC SUSCEPTIBILITY TESTING, ANTIMICROBIAL RESISTANCE PLASMID DNA, LIPASE
722
ARTICLE INFORMATION:
*Corresponding Author: farheena1980@gmail.com
Received 21
st
Oct, 2017
Accepted after revision 27
th
Dec, 2017
BBRC Print ISSN: 0974-6455
Online ISSN: 2321-4007 CODEN: USA BBRCBA
Thomson Reuters ISI ESC and Crossref Indexed Journal
NAAS Journal Score 2017: 4.31 Cosmos IF: 4.006
© A Society of Science and Nature Publication, 2017. All rights
reserved.
Online Contents Available at:
http//www.bbrc.in/
DOI: 10.21786/bbrc/10.4/17

Farheena, Chithira and Mohankumar
INTRODUCTION
Wound infection is one of the major health problems
resulting from colonization of wounds by pathogenic
organisms Recurrently the predominant Staphylococcus
aureus is one of the most important opportunistic patho-
gen among Staphylococci belonging to Micrococaceae
family causing signi cant infections under appropriate
conditions (Prescott et al., 2002). About 20%–30% of
the general populations are carriers of S. aureus. The
anterior nasal cavity is the main site of S. aureus car-
riage. Among nasal S. aureus carriers, approximately
one-half also carry the organism on their skin. Recent
studies have established that S. aureus is often found at
nonnasal sites, particularly the pharynx and the gastro-
intestinal tract, with some carriers having colonization
con ned to these sites (Mertz et al., 2009; Acton et al.,
2009, Fagbomedo and Femi-Ola, 2017).
Staphylococcus aureus is one of the most versatile
nosocomial (i.e. acquired in hospital) and dangerous
human pathogen since publication of its role in sepsis
by Ogston in 1880 and 1882 (Lowy, 1998). At present,
Staphylococcal resistance to antibiotic has been associ-
ated with resistant plasmids (R-plasmid) that have the
ability to mediate the production of drug inactivated
enzymes such as -lactamase.The spread of resistance
to antimicrobial agents in S. aureus is largely due to the
acquisition of plasmids and or transposons (Lyon and
Skurray, 1987Adeleke and Odelola, 1997; Adeleke et al.,
2002 and other functions (King et al., 2006 and Diep
et al., 2008).
The un-hygienic hand swab showed presence of MRSA
and improvement in hand hygiene, coinciding with a
reduction of nosocomial infections and MRSA transmis-
sion (Pittet et al., 2000). Treatment options for patients
with serious invasive infections caused by methicillin-
resistant Staphylococcus aureus (MRSA) are limited
(Drew, 2007).Increasing resistance of MRSA in recent
years has had a signi cant impact on several aspects
of patient care and infection control. Antibiotic policies
need to be updated regularly, along with comprehen-
sive monitoring of antibiotic prescribing and antibiotic
consumption in healthcare settings. These facts clearly
highlight the need of a characterization of MRSA strains
at a regular basis at all levels. With the increasing inci-
dence of MDR, recourse to new antibiotics has become
necessary.Microbial enzymes have many advantages
over the animal and plant enzyme, rstly; they are eco-
nomical and can be produced on large scale within the
limited space and time. Secondly, they are capable of
producing a wide variety of enzymes; they can grow a
wide range of environmental condition. Enzymes have
many roles in the pharmaceutical and diagnostic indus-
tries. Therefore the main objective of this study is to
isolate lipase enzyme from mangrove sediment bacteria
to treat against MRSA isolates.
MATERIAL AND METHODS
In the present study, 90 pus samples were collected from
various Hospitals in and around Tirupur District from
November 2012 to February 2013. The samples were
processed in the laboratory of PG and Research Depart-
ment of Zoology, CGAC. First step done was to isolate
the organisms from pus samples and then studied the
culture susceptibility of Staphylococcus aureus. All pus
samples were directly streaked on Mannitol Salt Agar
plates and incubated aerobically at 37ºC for 24 hours.
The isolates were identi ed with standard tests used to
identify S. aureus such as Gram stain, IMViC, Nitrate
reduction, Oxidase, catalase, slide and tube coagulase
tests. (Forbes et al., 2007).
Antibiotic sensitivity testing (AST) was done only for
con rmed S.aureus strains. Antibiotic sensitivity was
performed by Disc Diffusion Method (Bauer et al., 1966).
Overnight cultures of S.aureus in nutrient broth were
ooded over the surface of Mueller Hinton agar plates.
The Mueller Hinton agar plates were allowed to dry
before applying antibiotic disc (Baddour et al., 2006).
Commercially available antibiotic discs were obtained
from (Himedia Labs, Mumbai, India) were gently and
rmly placed on the agar plates, which were then left
at room temperature for 1 hour to allow diffusion of
the antibiotics into the agar medium. The plates were
then incubated at 37°C for 24 hours. If an antimicro-
bial activity was present on the plates, it was indicated
by an inhibition zone. The diameter of the inhibition
zones was measured in millimeter after 24 hours using a
standard chart and detected various pattern of antibiot-
ics as sensitive, intermediate and resistant to MRSA.
Multiple drug resistant strains of S. aureus which
showed 65% resistance to the antibiotics were selected
for plasmid extraction. Isolation of plasmids was per-
formed by Alkaline Lysis Method (Jegadeesh Babu and
Rajamanickam, 1998). The plasmids were observed in
0.7% Agarose gel electrophoresis.Extra cellular enzyme
such as lipase was detected from the marine bacteria
isolated from mangrove soil sediment. Using plate pre-
cipitation test with their speci c media screening of
lipolytic activity was done by using Rhodamine-B agar
plate (Kouker and Jaegar, 1987). The antibacterial activ-
ity of the lipase was performed by using well diffusion
method. So, hence in view to the signi cance of man-
grove ecosystem which provide a rich source of novel
lipase against MRSA isolates.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS ISOLATION AND IDENTIFICATION OF PHARMACEUTICALLY ACTIVE LIPASE PRODUCING BACILLUS SPP. 723
Farheena, Chithira and Mohankumar
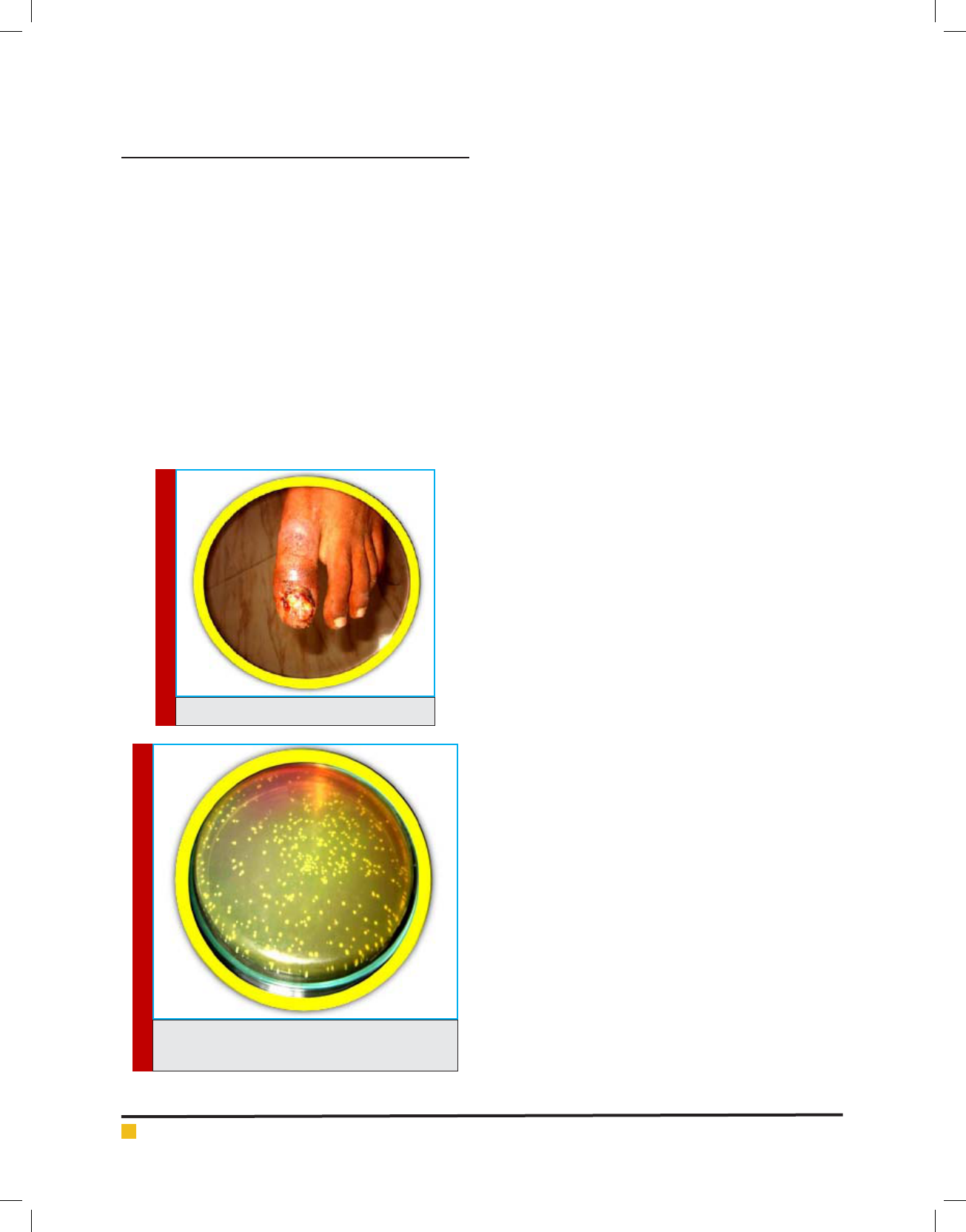
PLATE 1. Wound Sample
PLATE 2. Single colonies of S. aureus in MSA agar
plate
RESULTS AND DISCUSSION
Among the 92 clinical pus sample (Plate: 1) isolates of
S. aureus, 50 positive Staphylococcus aureus (Plate:
2) were identi ed as methicillin resistant Staphylococ-
cus aureus (MRSA) by disc diffusion method (Plate: 3).
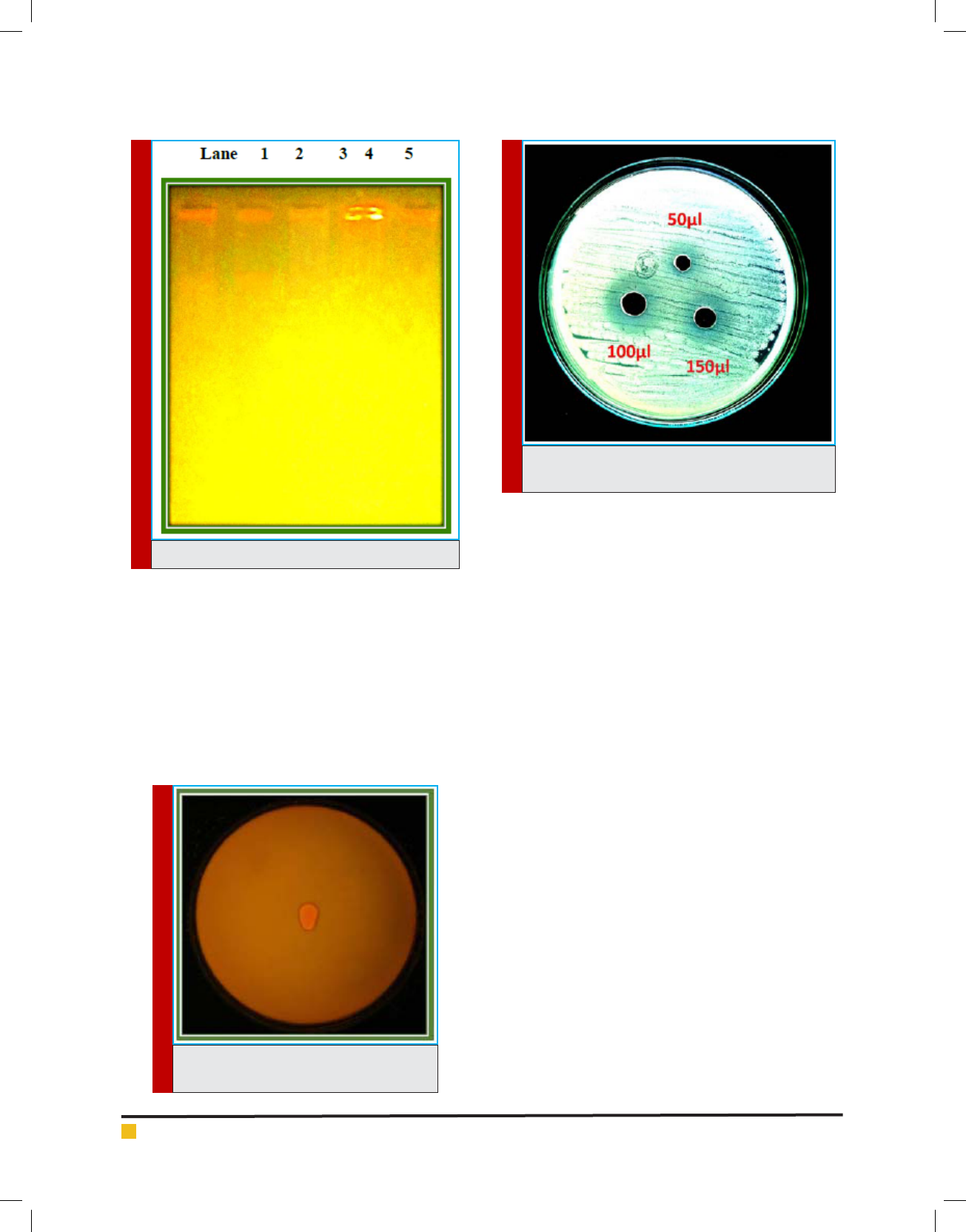
Electrophoretic analysis of the plasmid DNA prepared
was carried out by agarose gel electrophoresis on 0.7%.
Five MRSA isolates (FA21, FA24, FA48, FA49 and FA50)
which was showed more than 65% resistance against
tested antibiotics were selected for plasmid isolation
(Plate: 4).Strain no FA49 harbored a single plasmid
DNA on basis of electrophoretic mobility on agarose gel.
Other four strains (FA21, FA42, FA48 and FA50) har-
bored a double plasmid DNA. The molecular size of the
plasmid DNA was calculated to be 1500bp and 1000bp
respectively. 100bp DNA ladder (Medox biotech, Chen-
nai) was used as marker DNA.
Marine bacterium Bacillus spp. was isolated from
marine sediment collected from Pitchavaram mangrove.
This bacterium was identi ed by biochemical characters.
This bacterium was identi ed by using ZoBell medi a and
lipase activity screening by Rhodamine B agar media
(Plate: 5) followed by the antimicrobial activity of lipase
enzyme was done by well diffusion assay (Plate: 6).
In Rhodamine B agar the isolates Bacillus spp. showed
halos around the bacterial colonies under ultra violet
irradiation. Kouker and Jaeger (1987) detected a plate
assay for microbial lipase in a medium containing tria-
cylglycerol with addition of uorescent dye rhodamine
B. Substrate hydrolyses causes the formation of orange
uorescence halos around bacterial colonies visible
upon UV irradiation. The isolated strains were screened
for lipase production.
Staphylococcus aureus is recognized as an important
bacterial pathogen contributing towards hospital infec-
tion, globally. Despite the use of potent antibiotic still
high mortality exist in case of Staphylococcus aureus
infection. In the present study, antibiotic susceptibility
pattern was assessed for Gram-positive cocci from pus
and a high resistance was recorded against antibiotics
tested. Despite the numerous studies found that advent
of antibiotics, it was though that warrant for the treat-
ment of the S. aureus related infection got issued but
due to the development of antibiotic resistant gene in
the plasmid of S. aureus could depend itself in a much
secured manner. So, these facts clearly highlight the
need of a characterization of MRSA strains at a regu-
lar basis at all levels. With the increasing incidence of
MDR, recourse to new antibiotics has become neces-
sary. In recent year the microbial enzymes have many
roles in the pharmaceutical and diagnostic industries.
Therefore the main objective of this study is to isolate
lipase enzyme from mangrove sediment bacteria to treat
against MRSA isolates.
Adame et al (2010) collected 250 samples from
healthy humans, cattle, sheep and goats for the isola-
tion of S. aureus and they reported that the antimi-
crobial susceptibility test showed highly susceptible
to Ciproxcin (91.1%), Nor oxacin (90.2%), Rifampicin
(73.2%), Streptomycin (72.3%), Erythromycin (71.4%),
Norbactin (64.3%), but the isolates showed resistant to
Ceftazimide (7.1%), Cefotaxime (14.3%), and Ampiclox
(31.3%). Totally 92 wound samples were collected from
infected human, among 50 S. aureus were isolated, Pen-
icillin showed 100% resistant against the isolates, Ami-
kacin and tetracycline showed 6% and 4% resistance
respectively in the present study.
Susmita Bhattacharya et al (2013) have isolated 280
MRSA strains from 714 Staphylococcus aureus of vari-
ous clinical samples. Among 280 MRSA, 21 strains were
724 ISOLATION AND IDENTIFICATION OF PHARMACEUTICALLY ACTIVE LIPASE PRODUCING BACILLUS SPP. BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS

BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS ISOLATION AND IDENTIFICATION OF PHARMACEUTICALLY ACTIVE LIPASE PRODUCING BACILLUS SPP. 725
Farheena, Chithira and Mohankumar
PLATE 3. Antibiotic susceptibility test of S. aureus
found to be resistant to Vancomycin by disc diffusion
test. Similar method was used in the present inves-
tigation for antimicrobial susceptibility test. Among
50 MRSA strains, 36 strains were found to be resist-
ant to Vancomycin.High rates of resistance to Penicillin
among S.aureus have been observed since 1959, when
this frequency was recorded at 80% which have been
extended to amoxicillin and to ampicillin. Giarola et al
(2012) observed that the 90% resistance to Penicillin,
14% to rifampicin and 59% to azithromycin. Similarly
high rates of resistance was observed against Penicil-
lin (100%) but none of the isolates showed resistance
against rifampicin in this study.
All the isolates of S.aureus were multidrug resistant
and one isolate was pane resistant for all the tested drugs.
50 % resistance was observed in penicillin, methicillin,
polymyxin – B and Chloramphenicol 95.5%, 77.3%,
68.2% and 51.5% respectively (Alebachew et al 2009).
Similarly Penicillin and methicillin showed 100% and
64% resistance respectively, whereas none of the isolates
726 ISOLATION AND IDENTIFICATION OF PHARMACEUTICALLY ACTIVE LIPASE PRODUCING BACILLUS SPP. BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
Farheena, Chithira and Mohankumar
PLATE 4. Plasmid Pro le of S. aureus
PLATE 5. Screening of Lipase enzyme from
Bacillus spp.
PLATE 6. Antimicrobial activity of lipase against S.
aureus
showed resistance against chloramphenicol. Totally 23
antibiotic drugs were tested, among 23 antibiotics, 12
antibiotics were showed were than 50% resistance.
Twenty four Staphylococcus aureus isolates were
tested invitro to determine their antibiotic susceptibil-
ity patterns by antibiotic disc diffusion method. All the
isolates showed multiple antibiotic resistances to the
antibiotics tested. All the isolates were 100% resistant
to Ampicillin, Amoxicillin Nalidixic acid (87.5%), Cef-
triaxone (75%), Streptomycin (54.16%), Erythromycin,
tetracycline, Kanamycin and Neomycin (25%), Oxacil-
lin (41.6%), Tobramycin and there was no resistance
found to Chloramphenicol, Vancomycin, Nor oxacin,
Cipro oxacin and Rifampicin (AI-Hamdani and Hamad,
2012). Totally 50 Staphylococcus aureus isolates were
tested invitro by similar method. Here also all the iso-
lates were showed multiple antibiotic resistance to the
drugs tested. But Chloramphenicol and Rifampicin were
showed 100% resistant.
Udo and Sarlchoo (2010) have characterized four
MRSA isolates , during plasmid analysis, four isolates
had showed three plasmid patterns, Isolate K6482 con-
tained two plasmids (28 and 26 Kb), K6531 and K6533
each contained three plasmids (28, 21 and41 Kb) and
K6552 contained two plasmids (41 and 41 Kb). In this
present study ve strains which showed more than
65% resistance against all drug tested were taken for
plasmid isolation, Isolate FA49 contained one plasmid
(1.5Kb) and the isolates FA21, FA24, FA48 and FA50
each contained two plasmids (1.5kb and 1Kb).Herari et
al (2008) studied that the lipase production in an indig-
enous lipolytic Bacillus spp. in media containing tribu-
tyrin, tween 80 and rhodomine B- Olive oil. The statisti-
cal model was used to predict the optimum experimental
conditions for bacterial growth and lipase production.
Similarly in the present study Bacillus spp. was used
for lipase production, it was isolated from the sediment
of Pichavaram mangrove, lipase production was con-
rmed by Rhodomine B-olive oil plate.Mohankumar and
Tamilselvi (2012) have isolated marine bacteria includes
Bacillus spp. Pseudomonas spp. Staphylococcus spp. and
Vibrio spp. for lipase production. Amount of lipase was

BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS ISOLATION AND IDENTIFICATION OF PHARMACEUTICALLY ACTIVE LIPASE PRODUCING BACILLUS SPP. 727
Farheena, Chithira and Mohankumar
estimated by plate assay method and titration. Similar
method was followed in the present study that the Bacil-
lus spp. was isolated from mangrove sediment samples
and the lipase production was estimated only by plate
assay method.
CONCLUSION
The present study has indicated that MRSA strains were
found to be develop resistance day by day to the currently
used antibiotics. This situation needs some novel thera-
peutic drugs from marine environment for an alternative
therapy for treatment of wound causing MRSA isolates.
REFERENCES
A1- Hamdaniand, A., Hamad G. (2012). Study of plasmid pro-
le, susceptibility patterns of clinical Staphylococcus aureus
isolated from patients with otitis media in Basrah. Journal of
Basrah Researches (Sciences), 38 (1): 79-89.
Acton, D.S., Tempelmans Plat Sinnige M.J, Van Wamel W, De
Groot N,Van Belkum A. (2009). Intestinal carriage of Staphy-
lococcus aureus: how does its frequency compare with that of
nasal carriage and what is its clinical impact. Eur J Clin Micro-
biol Infect Dis. 28: 115 - 127.
Adamu, J.Y., Raufu A.I, Chimaroke F.C, Ameh J.A. (2010). Anti-
microbial susceptibility testing of Staphylococcus aureus iso-
lated from apparently healthy humans and animals in Maidu-
guri, Nigeria. International Journal of Biomedical and Health
Sciences, 6 (4): 191-195.
Adeleke, O.E., Odelola H.A, Oluwole F.A. (2002). Curing of anti-
biotic resistance in clinical strains of Staphylococcus aureus.
Afr J Med Pharm Sci. 6: 19-25.
Adeleke, O.E., Odelola H.A. (1997). Plasmid pro les of multiple
drug resistant local strains of Staphylococcus aureus. Afr J
Med Pharm Sci. 26: 119-121.
Alebachew, Gizachew Yismaw, Ayelegn Derabe, Zufan Sisay.
(2012). Staphylococcus aureus burn wound infection among
patients attending yekatit 12 hospital burn unit, ADDIS
ABABA, Ethiopia. Ethiop. J. Health. Sci, 22 (3) 209-213.
Baddour, M.M., Abuelkheir M.M., Fatani J.A. (2006). Trends in
antibiotic susceptibility patterns and epidemiology of MRSA
isolates from several hospitals in Riyadh, Saudi Arabia. Annals
of Clinical Microbiology and Antimicrobials, 5(30): 1- 11.
Bauer, A.W., Kirby W.M., Sherris J.C., Truck M. (1996). Antibi-
otic susceptibility testing by a standardized single disk method.
Am.J.Clin. Pathol., 45 (4): 493- 496.
Diep, B.A., Chambers H.F., Graber C.J, Szumewski J.D, Miller
L.G, Han L.L , Chen J.H, Lin F. (2008). Emergence of multidrug
resistant Staphylococcus aureus clone USA 300 in man who
have sex with men. Ann Intern Med.148: 1-17.
Drew, R.H. (2007). Emerging options for treatment of invasive
multidrug resistant Staphylococcus aureus infections Pharma-
cotherapy, 27: 227-49.
Edet Udo, E., Eiman Sarkhoo.(2010). Genetic analysis of high
level mupirocin resistance in the ST80 clone of community
associated methicillin resistant Staphylococcus aureus. Journal
of Medical Microbiology, 59: 193-199.
Fagbomedol J and Femi-Ola, T.O (2017). Incidence of wound
infections and the prevelance of multi drug resistant Staphy-
lococcus aureus in Nigerian hospital. Academia Journal of Sci-
enti c Research. 5(9): 316-322.
Giarola, Rosiane Ribeiro dos Santos, Maria Cristina Bronharo
Tognim, Sueli Donizete Borelli, Joao Bedendo. (2012). Carriage
frequency, phenotypic and Genotypic characteristics of Staph-
ylocoous aureus isolated from dialysis and kidney transplant
patients at a hospital in Northern Parana. Brazilian journal of
Microbiology, 923-930.
Herari, K., Eftekhar F, Yakhchali B, Tabendeh F. (2008). Isola-
tion and identi cation of lipase producing Bacillus spp from
soil. Pak. J. Bio. Sci, 11: 740-745.
King, M.D., Humphrey B.J, Wang Y.F. (2006). Kourbalova EV,
Ray SM , Blumbrg HM; Emergence of community acquired
methicillin resistant Staphylococcus aureus USA 300 clone as
the predominant cause of skin and soft tissue infections. Ann
Intern Med. 144: 309-317.
Kouker, G., Jaeger K.E. (1987). Speci c and sensitive plate
assay for bacterial lipases. App.Environ. Microbiol. 53: 211-
213.
Lowey, F.D. (1998). Staphylococcus aureus infections. New Eng
J Med. 339: 520- 532.
Lyon, B.R, Skurray R. (1987). Antimicrobial resistance of
Staphylococcus aureus: genetic basis. Micr Rev. 51: 88-134.
Mertz, D., Frei R, Periat N. (2009). Exclusive Staphylococcus
aureus throat carriage: at risk populations. Arch Intern Med.
69: 172-178.
Mohankumar, A., Tamilselvi S. (2012). Antibiotic pattern of
met hicillin resistant
Staphylococcus aureus is chronic wound
of isolated from chronic wound of sherman community.
International Journal of Microbiological Research, 3 (2): 109-
116.
Pittet, D.S., Hugonnet S.H, Mourouga P, Touveneau S, Per-
neger T.V. (2000). Effectiveness of hospital wide programme
to improve compliance with hand hygiene. The Lancet, 356:
1307-1312.
Prescott, L.M., Harely J.P, Klein D.A. (2002). Microbiology 5
th
McGraw Hill Comp. USA.
Susmita Bhattachrya, Kuhu Pal, Mitali Chatterjee, Mandira
Banerjee, Pratip Kumar Kundu, Swapan Kumar Niyogi. (2013).
Vancomycin intermediate Staphylococcus aureus isolated from
a tertiary care hospital in Kolkata. IOSR Journal of Dental and
Medical Sciences, 5 (2): 19-23.