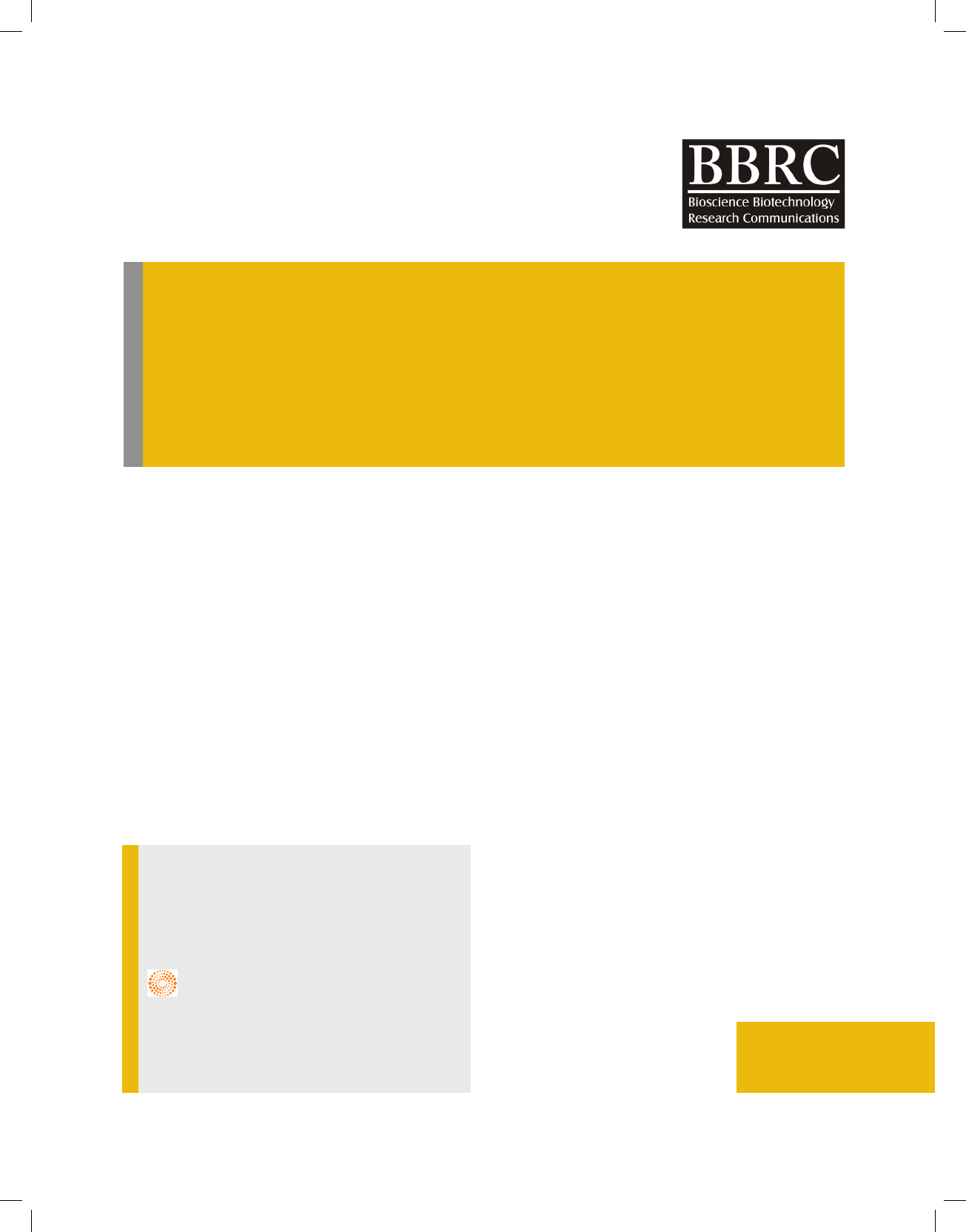
Isolation, biochemical characterization and antibiotic
pro ling of members of
Enterobacteriacea
isolated from
animal fecal matter
Azra Rashid
Department of Microbiology, Sardar Bhagwan Singh PG Institute of Biomedical Sciences and Research,
Af liated to HNB Garwal University, Dehradun, India
ABSTRACT
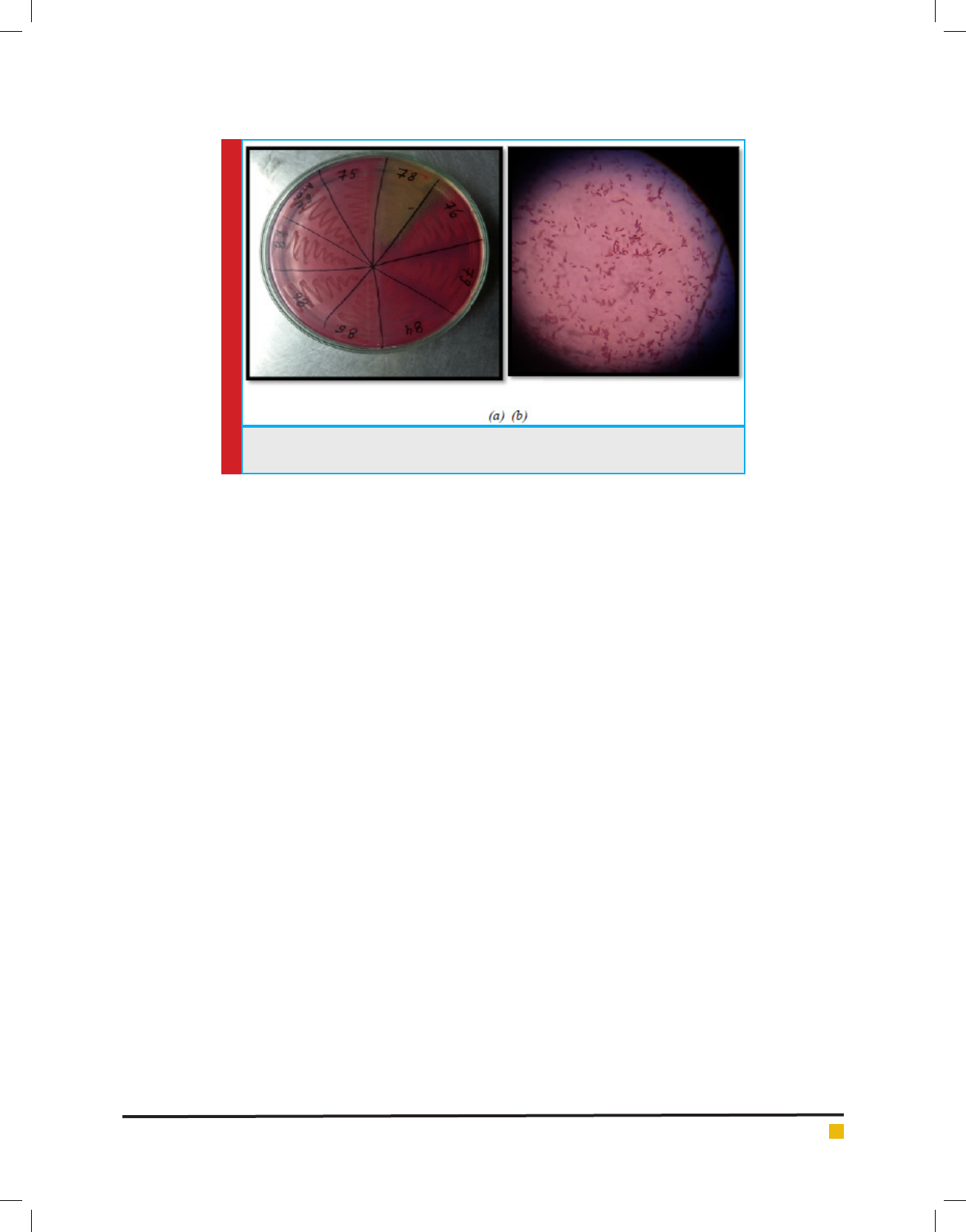
In the present study, fecal samples were collected from different localities of Balawala, Dehradun city. Out of 87 iso-
lates, 50% were E. coli, 25% were Klebsiella spp., 15% were Enterobacteriaceae spp. and 10% were Proteus spp. The
isolates were then checked for antibiotic sensitivity. 50% strains were resistant for novabiocin, 25% were resistant for
ce xime, 15% were resistant for clotrimazole and 10% were resistant for amoxicillin and most of these showed sensitiv-
ity against the antibiotics- Amikacin, amoxicillin, ce xime, cephalexin, cipro oxacin, clotrimazole, gentamicin, nova-
biocin, o oxacin and trimethoprim. In the minimum inhibitory concentration test, 50% of the isolates showed resist-
ance against the antibiotics amoxicillin, ampicillin, streptomycin at different concentrations (8µg/ml, 16µg/ml, 32µg/
ml, 64µg/ml and 128µg/ml respectuvely) and 50% showed sensitivity against the antibiotics cefoparazone sulbactum,
meropenem and piperacillin tazobactum. In conclusion, the data of the present study determine the resistance pro le of
enteric pathogens in animal fecal samples and is helpful from the community infection point of view. The study provides
some insight on the prevalence dynamics of enteric pathogens from animal fecal which can be helpful to clinicians to
formulate proper antimicrobial therapy.
KEY WORDS: AMIKACIN, RESISTANCE PROFILE, MINNIMUM INHIBITORY CONCENTRATION, MRSA, ENTEROBACTERIACEA
603
Microbiological
Communication
Biosci. Biotech. Res. Comm. 11(4): 603-611 (2018)
ARTICLE INFORMATION:
Corresponding Authors: publication.cytogene@gmail.com
Received 17
th
Sep, 2018
Accepted after revision 19
rd
Dec, 2018
BBRC Print ISSN: 0974-6455
Online ISSN: 2321-4007 CODEN: USA BBRCBA
Thomson Reuters ISI ESC / Clarivate Analytics USA
Mono of Clarivate Analytics and Crossref Indexed
Journal Mono of CR
NAAS Journal Score 2018: 4.31 SJIF 2017: 4.196
© A Society of Science and Nature Publication, Bhopal India
2018. All rights reserved.
Online Contents Available at:
http//www.bbrc.in/
DOI: 10.21786/bbrc/11.4/10

Azra Rashid
604 ISOLATION, BIOCHEMICAL CHARACTERIZATION AND ANTIBIOTIC PROFILING OF MEMBERS BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
INTRODUCTION
Bacterial resistance to antibiotics continues to curb our
ability to treat, cure and control infectious diseases. Two
organisms in particular that have become major public
health threats are methicillin-resistant Staphylococcus
aureus and penicillin-resistant Streptococcus pneumo-
niae. Resistance to aminocyclitol aminoglycosides is an
important clinical problem since these antibiotics are
widely used in the treatment of serious infections, (Lar-
son et al., 1986; Garcia et al., 1989). Large quantities of
enteric bacteria from animal fecal wastes can be released
into rivers and lakes that serve as sources of water for
drinking, recreation or irrigation. Fecal contamination is
considered to be main contributor of enteric pathogens
to natural water sources. Infection originating from such
sources specially diarrhea and typhoid fever. The family
of Enetrobacteriaceae is accountable for these illnesses.
The important members of Enterobacteriaceae are E. coli,
Salmonella and Shigella. Amikacin has been the drug
of choice for treating nosocomial infections refractory
to other aminoglycosides (Gerding et al., 1990; Levine
et al., 1985, Kalita et al., 2016).
In recent years, resistance to amikacin due to pro-
duction of 3’-aminoglycoside-phosphotransferases, 2”
–adenyltransferases and aminoglycoside-6’- N-acetyl-
transferases has been reported (Hopkins et al., 1991;
Shaw et al., 1993; Shimizu et al., 1985). Transmis-
sion of this microbe is usually through uncooked meats
and eggs. The disease is spread via the fecal-oral route
and requires very low cell numbers to initiate infection.
In many cases, Shigella infection will lead to diarrhea
accompanied by fever. Among the disease caused by
poultry and other farms and their products some are
often severe and sometimes lethal infection such as
meningitis, endocarditis, urinary tract infections, septec-
imia, epidemic diarrhea of adults and children. Resist-
ance are more commonly observed among isolates of
animal fecal. The relatively intensive conditions under
which animal are housed may be associated with greater
disease potential and therefore a greater potential and
therefore a greater tendency for antibiotic use of disease
control (Bywater et al., 2004).
Resistance to antimicrobials and particularly mul-
tidrug resistance is an emerging problem in Entero-
bacteriaceae for developing and developed countries
(Schwarz and White, 2005). Resistant microorganisms
have emerged as a result of improper use of antibiot-
ics in human health as well as in agricultural practices
(Khachatourians, 1998). Investigators have reported
evidence of some low-level resistance to antibiotics,
but overall the bacteria studied were sensitive to most
antibiotics prior to exposure (Datta and Hughes, 1983;
Dancer, 1997).
MATERIALS AND METHODS
Isolation of Enteric Pathogens: Sample was diluted
appropriately in sterile saline by serial dilution method
and then an appropriate dilution (0.1ml) was plated on
selective media and incubated at 37 ˚C for 24 to 48 h
(Pelcezar et al., 1986) and then observed for the growth.
Identi cation and characterization of Enteric patho-
gens: All suspected colonies on respective selective
media were presumptive forms identi ed using identi-
cation scheme of Bergey’s manual (1997) that identi-
es bacteria on the basis of morphological, cultural and
biochemical characteristics. The methods suggested in
the microbiological methods were followed (Borrego
and Figueras, 1997) for characterization of the bacte-
rial isolates.
Antibiotic Susceptibility Test: Bacterial isolates viz.,
E. coli, Enterobacteriaceae, Klebsiella sps., Proteus sps.
were screened for their sensitivity to antibiotics because
the frequency of occurrence of these pathogens was
very high. Multidrug resistant strains of these pathogens
are emerging worldwide. Overnight growth of respec-
tive bacterial isolates was used for the sensitivity test.
The Kirby Bauer modi ed disk diffusion technique was
was used to determine the sensitivitity to antibiotics.The
polydiscs (Micromaster Laboratories) were evenly dis-
tributed on sterile Mueller Hinton agar medium. Plates
were then incubated at 37 ˚C for 24 h. The inhibition
zone diameters were measured using meter scale. Inhibi-
tion zone diameters were compared with the standard
inhibition zone for resistance, intermediate and suscep-
tible character (Kalita et al., 2016).
Minimal Inhibitory Concentration (MIC): Minimum
inhibitory concentration was determined according to
the method described earlier by adding various concen-
trations of antibiotics (8-128 g/ml) in Nutrient Broth.
Further, 100 l of inoculum was added to each tube and
incubated the tubes at 37°C for 24 hours (Sharma et al.,
2011).
RESULTS AND DISCUSSION
Isolation of Enteric pathogens from Animal excreta:
Samples of animals were collected aseptically and trans-
ported to the laboratory immediately for isolation of
enteric pathogens on Mac-Conkey agar, Eosine meth-
ylene agar, Cystine–lactose–electrolyte-de cient agar
plates. The plates were incubated for 14- 16 hours at
37°C and after incubation observations were made there
are appearances of isolated colonies. The isolated colo-
nies were further pure cultured by sub-streaking on
Mac-Conkey agar plates (shown in Fig. 1). The culture
Azra Rashid
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS ISOLATION, BIOCHEMICAL CHARACTERIZATION AND ANTIBIOTIC PROFILING OF MEMBERS 605
FIGURE 1. Isolation of Enteric pathogen(a) Selective Isolation of Enteric Pathogens on
Ma Conkey Agar (b)Microscopic Observation- Gram Negative Rods
thus obtained and the details of healthy animals and
humans are given in Table 1
In this study susceptibility pattern of pathogens liable
for urinary tract infections in Poland to cogently used
antimicrobial agents. A most entire study of 141 patho-
gens from hospital – acquired infections and 460 patho-
gens from community- acquired infections were isolated
between July 1998 and May 1999. The most common
ecological agent was E. coli (73.0 %), followed by Pro-
teus spp. (8.9 %) and other species of Enterobacteriaceae
(9.6 %). Few community infections were caused by
Gram-positive cocci were isolated more frequently from
a hospital setting (14.1 %) and the most common was
Enterococcus spp. (8.5 %). Pseudomonas aeruginosa was
found only among hospital isolates and was responsible
for 10.7 % of infections. E.coli isolates from both com-
munity and hospital infections were highly affected to
many antimicrobial agents with the explusion of those
isolates generating elongated spectrum beta- lactamases
(ESBLs). Of all Enterobacteriaceae tested, 38 strains (6.9
%) were able to generating ESBLs (Ahmed et al., 2011).
ANTIBIOTIC SENSITIVITY TEST
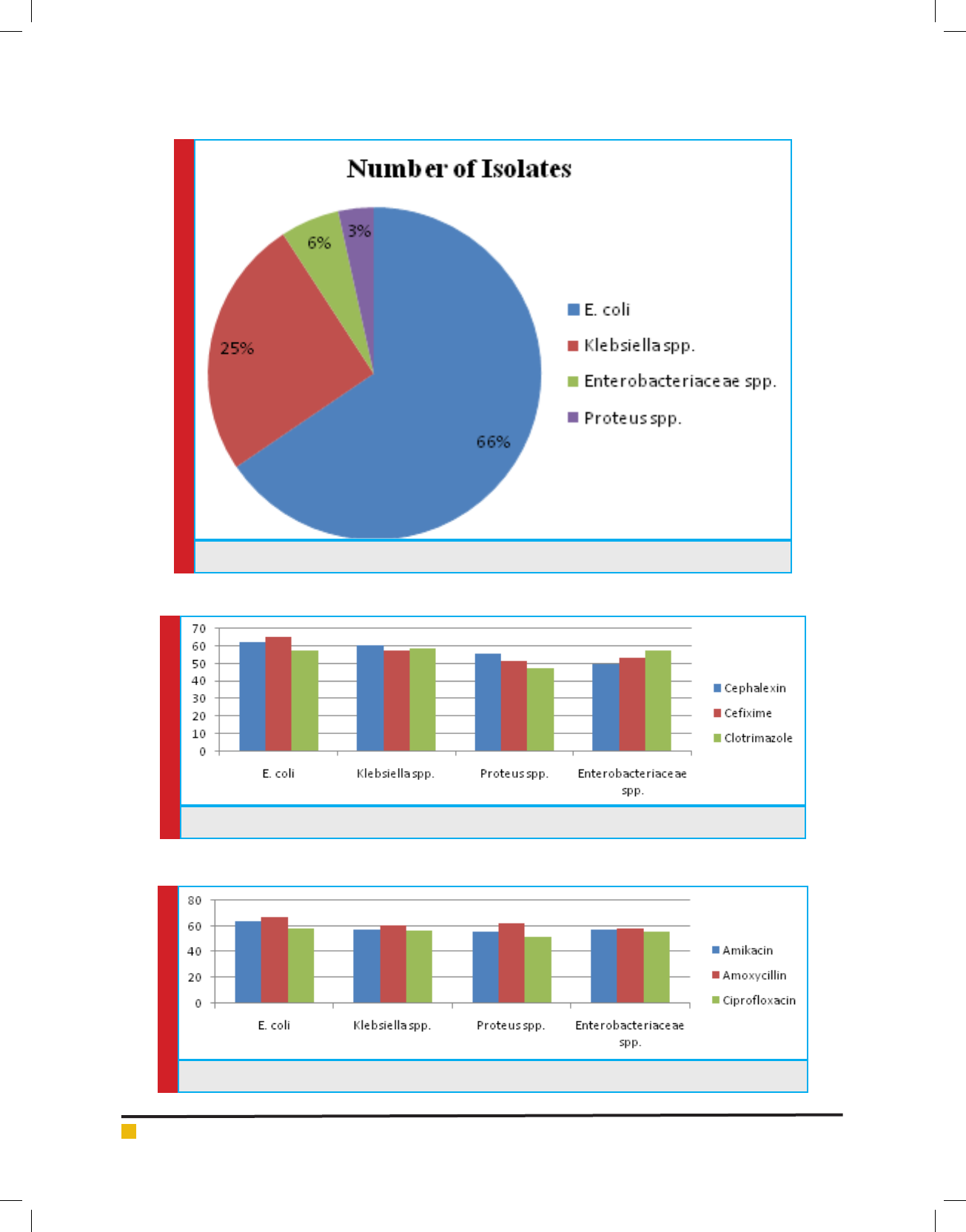
Antibiotic sensitivity of all the 87 isolates was deter-
mined against 10 antibiotics belonging to -lactam and
non -lactam group. The antibiotics included are Ami-
kacin, Amoxycillin, Ce xime, Cephalexin, Cipro oxa-
cin, Clotrimazole, Gentamycin, Kanamycin, Novabiocin
and O oxacin. There sensitivity to different antibiotics
is represented in Graph 1, 2 & 3.
According to Ergin & Mutlu, 197 bacterial isolates
from Sudanese patients with diarrhea or urinary tract
infections. Shigella dysenteriae type 1 and enteropatho-
genic E. coli Showed high resistance rates against the
commonly used antimicrobial agents: ampicillin, chlo-
ramphenocol, amoxycillin, co-trimoxazole, tetracy-
cline, malidixic acid, sulfonamide and neomycin. The
uropathogens wre completely sensitive to cipro oxa-
cin. Resistance to tetracycline, amoxicillin, ampicillin,
cotrimoxazole and sulfonamide was the most frequent
pattern. The common urinary tract pathogens Klebsiella
pneumonia, E. coli and Proteus mirabilis showed high
rates of resistance to ampicilin, co-trimoxazole, amoxi-
cillin, tetracycline, trimethoprim, sulfonamide, strepto-
mycin and carbenicillin.
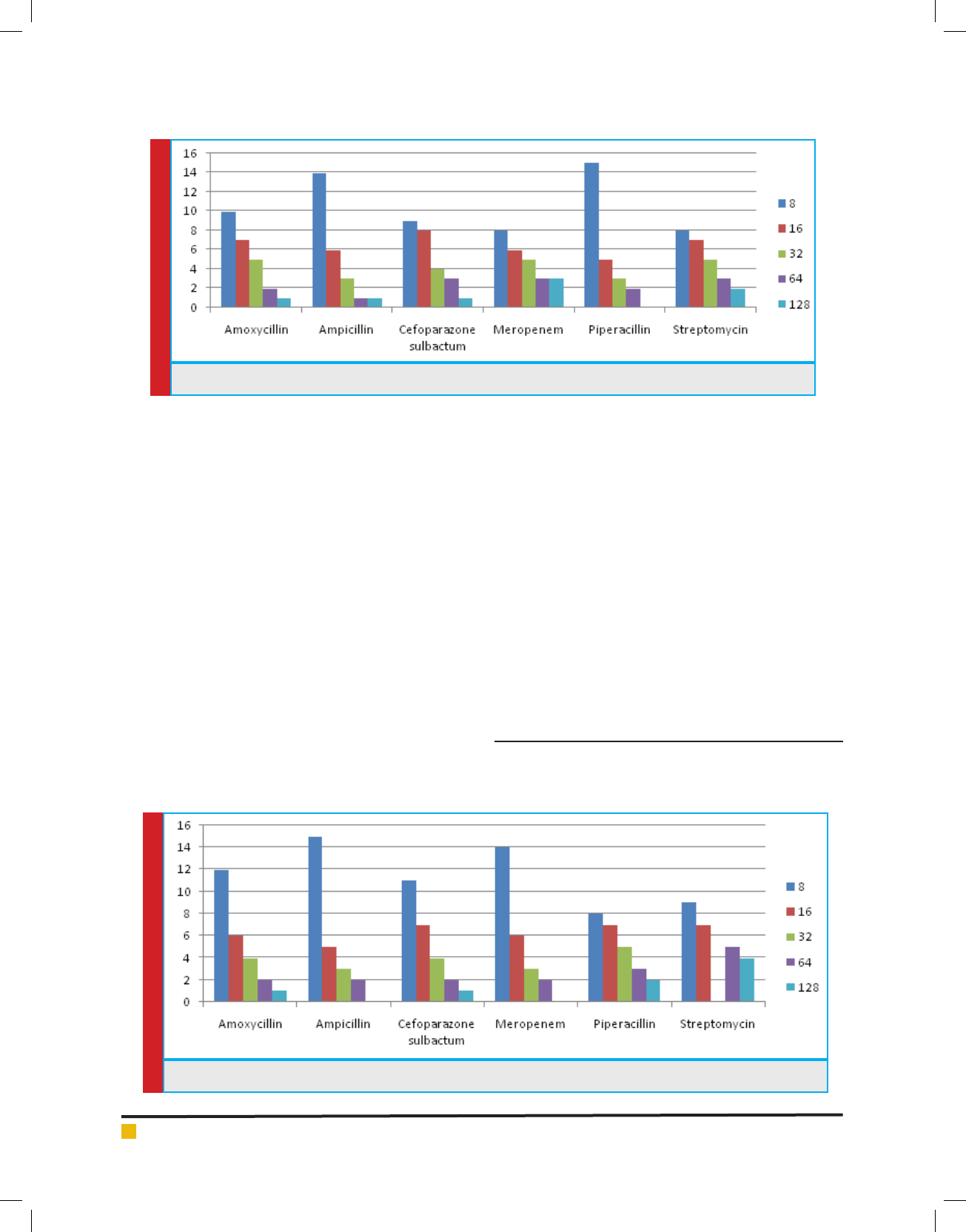
MINIMUM INHIBITORY CONCENTRATION
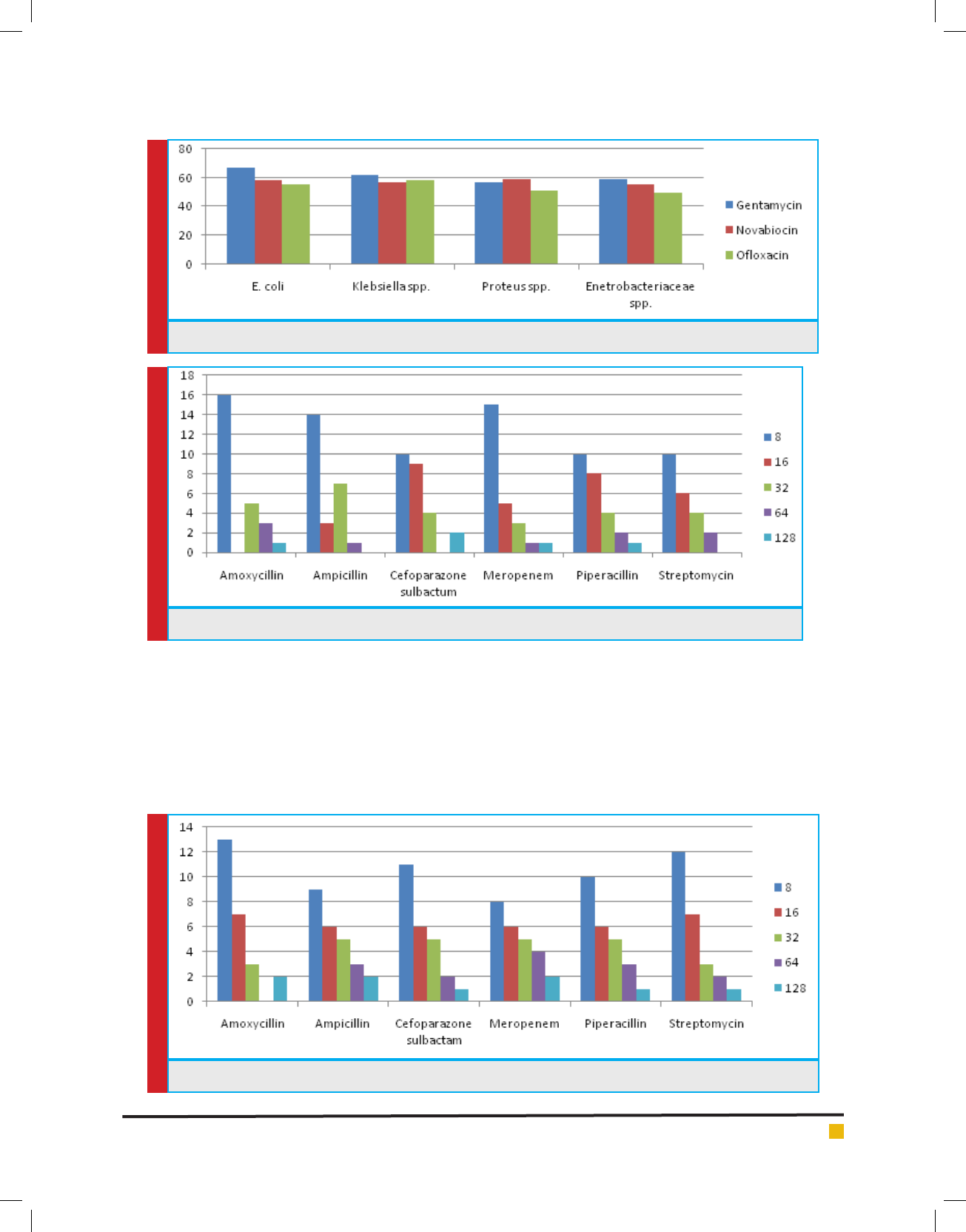
Of all 87 samples 25 samples were selected for carry-
ing out MIC of Amoxycillin, Ampicillin, Pipracillin
tazobactum, Streptomycin, Meropenem and Cefopara-
zone sulbactum. The MIC was conducted at different
concentrations like (8µg, 16µg, 32µg, 64µg and 128µg).
Maximum isolates showed resistance against Amoxycil-
lin and minimum against Meropenem and Cefoparazone
sulbactum. In decresing order of resistance antibiotics
can be placed as Amoxycillin>Ampicillin>Streptomycin
>Pipracillintazobactum>Meropenem>Cefoparazone sul-
bactum. The MIC result of isolates is shown in Graph.
4, 5, 6 & 7.
In this study they determined the distribution rates of
Pseudomonas aeuroginosa in clinics and its resistance to
antibiotics. The antibiotic resistance rates were detected
by minimal inhibitory concentration (MIC). The clinical
and specimen distribution properties of Pseudomonas
were evaluated based on their resistance pattern. Pseu-
domonas was the fourth common bacteria in all isolates.

Azra Rashid
606 ISOLATION, BIOCHEMICAL CHARACTERIZATION AND ANTIBIOTIC PROFILING OF MEMBERS BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
Table 1. Cultures obtained from Animal Fecal Matter
S. No Sample Number Growth On MacConkey Agar Morphology Motility
1. AH1 Small pink color colonies/ pink background Small pink color colonies and mucoid +ve
2. AH2 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
3. AH3 Pink color colnies/ pink background Small pink color colonies and mucoid -ve
4. AH4 Pink color colonies/ pink background Small pink color colonies and mucoid -ve
5. AH5 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
6. AH6 Pink color colonies/ pink background Small pink color colonies and mucoid -ve
7. AH7 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
8. AH8 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
9. AH9 Pink color colonies/ pink background Small pink color colonies and mucoid -ve
10. AH10 Yellow swarming colonies/ yellow background Yellow color colony and show motility +ve
11. AH11 Small pink color colonies/ pink background Small pink color colonies and mucoid +ve
12. AH12 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
13. AH13 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
14. AH14 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
15. AH15 Yellow color colonies/ yellow background Gram –ve, non-motile and rod shaped -ve
16. AH16 Colorless colonies/ white background Gram –ve, non-motile and rod shaped +ve
17. AH17 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
18. AH18 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
19. AH19 Translucent gummy colonies/ pink background Gram –ve, non-motile and rod shaped +ve
20. AH20 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped +ve
21. AH21 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
22. AH22 Pink color colonies/ pink background Small pink color colonies and mucoid -ve
23. AH23 Translucent gummy colonies/ pink background Gram –ve, non-motile and rod shaped +ve
24. AH24 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped +ve
25. AH25 Yellowish gummy colonies/ yellow background Gram –ve, non-motile and rod shaped -ve
26. AH26 Colorless colonies/ white background Gram –ve, non-motile and rod shaped +ve
27. AH27 Colorless colonies/ white background Gram –ve, non-motile and rod shaped +ve
28. AH28 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
29. AH29 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
30. AH30 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
31. AH31 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
32. AH32 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
33. AH33 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
34. AH34 Pink color colonies/ pink background Small pink color colonies and mucoid -ve
35. AH35 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped +ve
36. AH36 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
37. AH37 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
38. AH38 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
39. AH39 Pink color colonies/ pink background Small pink color colonies and mucoid -ve
40. AH40 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
41. AH41 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
42. AH42 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
43. AH43 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped +ve
44. AH44 Pink color colonies/ pink background Small pink color colonies and mucoid +ve

Azra Rashid
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS ISOLATION, BIOCHEMICAL CHARACTERIZATION AND ANTIBIOTIC PROFILING OF MEMBERS 607
44. AH44 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
45. AH45 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped +ve
46. AH46 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped -ve
47. AH47 Translucent gummy colonies/ pink background Gram –ve, non-motile and rod shaped +ve
48. AH48 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
49. AH49 Small orange color colonies/ pink background Gram –ve, non-motile and rod shaped +ve
50. AH50 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
51. AH51 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped +ve
52. AH52 Pink color colonies/ pink background Small pink color colonies and mucoid -ve
53. AH53 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
54. AH54 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
55. AH55 Translucent gummy colonies/ pink background Gram –ve, non-motile and rod shaped +ve
56. AH56 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
57. AH57 Translucent gummy colonies/ pink background Gram –ve, non-motile and rod shaped +ve
58. AH58 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped +ve
59. AH59 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
60. AH60 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped -ve
61. AH61 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
62. AH62 Yellow swarming colonies/ yellow background Yellow color colony and show motility +ve
63. AH63 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
64. AH64 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped -ve
65. AH65 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
66. AH66 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
67. AH67 Yellow swarming colonies/ yellow background Yellow color colony and show motility +ve
68. AH68 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
69. AH69 Colorless colonies/ pink background Gram –ve, non-motile and rod shaped +ve
70. AH70 Translucent gummy colonies/ white background Gram –ve, non-motile and rod shaped +ve
71. AH71 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
72. AH72 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
73. AH73 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
74. AH74 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
75. AH75 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
76. AH76 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
77. AH77 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
78. AH78 Translucent gummy colonies/ yellow background Gram –ve, non-motile and rod shaped +ve
79. AH79 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
80. AH80 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
81. AH81 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
82. AH82 Pink color colonies/ white background Small pink color colonies and mucoid +ve
83. AH83 Colorless colonies/ white background Gram –ve, non-motile and rod shaped +ve
84. AH84 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
85. AH85 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
86. AH86 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
87. AH87 Pink color colonies/ pink background Small pink color colonies and mucoid +ve
Azra Rashid
608 ISOLATION, BIOCHEMICAL CHARACTERIZATION AND ANTIBIOTIC PROFILING OF MEMBERS BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
FIGURE 2. Cultures obtained from Animal Fecal Matter
GRAPH 1. Antibiotic Sensitivity Of Selected Strain To -Lactam Antibiotics
GRAPH 2. Antibiotic Sensitivity Of Selected Strain To Non -Lactam Antibiotics
Azra Rashid
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS ISOLATION, BIOCHEMICAL CHARACTERIZATION AND ANTIBIOTIC PROFILING OF MEMBERS 609
GRAPH 3. Antibiotic Sensitivity Of Selected Strain To Non -Lactam Antibiotics
GRAPH 4. MIC Concentration Against Different Antibiotics for E.Coli
GRAPH 5. MIC Concentration Against Different Antibiotics for Enterobacteriaceae sps.
Tracheal aspirates, sputum and wound, pus were impor-
tant sources for Pseudomonas aeruginosa isolation in
intensive and nonintensive care units of surgery wards
(SW-ICU, SW-nonICU) (p<0.05). on the basis of MIC cri-
teria, the resistance ratios of the isolates to cefriaxone,
cefotaxime, ceftazidime, imipenem, o oxacin and cip-
ro oxacin were 8.4%, 15.0%, 13.3%, 0.0%, 11.6 % and
8.3% respectively (Hryniewicz et al., 2001).
A wide range of pathogenic microorganisms can be
transmitted to humans via water contaminated with
fecal matter. These include enteropathogenic agents
such as E. coli, Shigella, salmonella, enteroviruses and
multicellular parasites as well as opportunistic patho-
gens like Pseudomonas aeruginosa, Klebsiella etc. Appli-
cations of antibiotics bring about an increase in resist-
ance to antibiotics not only in pathogenic bacterial
Azra Rashid
610 ISOLATION, BIOCHEMICAL CHARACTERIZATION AND ANTIBIOTIC PROFILING OF MEMBERS BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
GRAPH 6. MIC Concentration Against Different Antibiotics for Klebsiella sps.
GRAPH 7. MIC Concentration Against Different Antibiotics for Proteus sps.
strains, but also in commensal bacteria (Luzzaro et al.,
2001)
In the present study, the samples from different local-
ities of Balawala were collected and total of 87 isolates
were obtained from them and among these 50% were E.
coli, 25% were Klebsiella spp., 15% were Enterobacte-
riaceae spp., and 10% were Proteus spp. The isolates were
then identi ed on the basis of biochemical characteris-
tics and the Klebsiella, E. coli, Proteus, Enterobacteria
and Pseudomonas were isolated from the excreta of ani-
mals. Antibiotic resistance among the isolates was also
evaluated using for antibiotics- amikacin, gentamicin,
novabiocin, o oxacin, cipro oxacin, cephalexin, ce x-
ime, amoxicillin, clotrimazole, trimethoprim, kanamy-
cin, ampicillin, streptomycin, meropenem, piperacillin
tazobactam and cefoparazone sulbactum.
In our study it has been seen that resistance was
seen for novabiocin (50%), ce xime (25%), clotrima-
zole (15%) and amoxicillin (10%). It was also found to
be sensitive for gentamicin, amikacin, kanamicin, tri-
methoprim, cipro oxacin and o oxacin. The MIC test
was also conducted during this study those isolates are
chosen for the MIC that showed more resistance ef -
cacy. The MIC has been performed by chosing the dif-
ferent isolates in which following antibiotics was used
viz amoxicillin, ampicillin, cefoparazone sulbactum,
meropenem, piperacillin tazobactum and streptomycin.
50% of the isolates showed resistance among the anti-
biotic amoxicillin, ampicillin, streptomycin at different
concentrations (8µg/ml, 16µg/ml, 32µg/ml, 64µg/ml and
128µg/ml) and 50% showed sensitivity against the anti-
biotic cefoparazone sulbactum, meropenem and pipera-
cillin tazobactum. The high density of enteric pathogen
and prevalence of multidrug resistant E. coli, Proteus
and Kleibsiella in the fecal matter may pose severe pub-
lic health risk.
CONCLUSION
In this study, we analysed the susceptibility pattern of
different aminoglycosides in different locality of Bala-

Azra Rashid
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS ISOLATION, BIOCHEMICAL CHARACTERIZATION AND ANTIBIOTIC PROFILING OF MEMBERS 611
wala, strain collections of E. coli, Klebsiella spp., Pseu-
domonas spp., Enterobacteriaceae spp. and Proteus spp.
Enteric pathogens, which are of great concern since they
are the most common causes of infection among humans
and animals. Aminoglycosides represent an important
class of antimicrobial agents. The prevalence of amino-
glycoside resistance among Gram-negative bacteria in
Dehradun is low, but an increased prevalence among
clinical isolates of Escherichia coli has been observed
during the last years. The most prevalent resistance
mechanism is aminoglycoside modifying enzymes.
REFERENCES
Ahmed AA, Osman H, MAnsour AM, Musa HA, Ahmed AB,
Karror Z, Hassan HS. (2000) Antimicrobial agent resistance in
bacterial isolates from patients and diarrhea and urinary tract
infection in the Sudan; Am J Trop Med Hyg.; 63 (5-6): 259-63.
Borrego JJ, Figueras MJ (1997) Microbiological quality of nat-
ural water s. Microbiologia 13:413-426
Bywater R, Deleryker H, Deroover E, Annode Jong, Mrion A,
McConville M, Rowan T and Walters J (2004) A European sur-
vey of the antimicrobial susceptibility among zoonotic and
commensal bacteria isolated from food producing animals, J.
Antimicrob. Chemotherap. 54: 744-754.
Dancer SJ, et al. (1997) Isolation and characterization of coli-
forms from glacial ice and water in Canada’s High Arctic. J
Appl Bacteriol; 82: 597-609.
Datta N, Hughes VM (1983) Plasmids of the same Inc groups in
enterobacteria before and after the medical use of antibiotics.
Nature; 306: 616-627.
Ergin C & Mutlu G. (1999) Clinical distribution and antibiotic
resistance of Pseudomonas sps. Eastern journal of medicine 4
(2); 65-69.
Garcia DC, Trevisan AR, Botto L, Cervetto M, Sarubbi MA,
Zorzopulos J (1989) An outbreak of multiply resistant Pseu-
domonas aeruginosa in a neonatal unit: plasmid pattern analy-
sis. J Hosp. Infect; 14: 99-105.
Gerding DN, Larson TA, Hughes RA, Weiler M, Shanholtzer C,
Peterson LR (1990) Aminoglycoside resistance and aminogly-
coside usage: ten years of experience in one hospital. Antimi-
crob Agents Chemother; 35: 1284-1290.
Hopkins JD, Flores A, Pilar-Pla M, Lester S, O’Brien TF (1991)
Nosocomial spread of an amikacin resistance gene on both a
mobilized, nonconjugative plasmid and a conjugative plasmid.
Antimicrob Agents Chemother 35: 1605-1611.
Hryniewicz K, Szczypa K, Sulikowski K, Bettejewska K, Hrynie-
wicz W (2001) Antibiotic susceptibility of bacterial strains iso-
lated from urinary tract infections in Poland, J Antimicrob
Chemother; 47(6): 773-80.
Kalita, S, Kandimalla, R, Sharma, KK, Kataki, AC, Deka, M and
Kotoky, A (2016) Amoxicillin functionalized gold nanoparti-
cles reverts MRSA resistance. Mater. Sci. Eng. C 61 720–727.
Khachatourians, G (1998) Agricultural use of antibiotics and
the evolution and transfer of antibiotic-resistant bacteria.
Canadian Medical Asso. J. 159: 1129-1136.
Larson TA, Garrett CR, Gerding DN (1986) Frequency of ami-
noglycoside 6’-N-acetyltransferase among Serratia species
during increased use of amikacin in the hospital. Antimicrob
Agents Chemother; 30: 176-178.
Levine JF, Maslow MJ, Leibowitz RE et al. (1985) Amikacin-
resistant Gram-negative bacilli: correlation of occurrence with
amikacin use. J Infect Dis; 151: 295-300.
Luzzaro F, Perilli M, Amicosante G, et al. (2001) Properties
of multidrug-resistant, ESBL-producing Proteus mirabilis iso-
lates and possible role of beta-lactam/beta-lactamase inhibitor
combinations. Int J Antimicrob Agents; 17 (2): 131-135.
Pelcezar MJ, Chan ECS, Krieg NR (1986) In: Microbiology. 5 th
edition. Tata McGraw- Hill Publishing Company pp 598-614.
Schwarz, S and White, D (2005) Phenolic resistance. Frontiers
in Antimicrob. Resistance. ASM press. 124-148.
Sharma, KK, Saikia, R, Kotoky, J, Kalita, JC and Das, J (2011)
Evaluation of antidermatophytic activity of piper beetle, Alla-
manda cathertica and their combination: an in vitro and in
vivo study. Int. J. PharmTech. Res. 3 644–651.
Shaw KJ, Rather PN, Hare RS, Miller GH (1993) Molecular
genetics of aminoglycoside resistance genes and familial rela-
tionships of the aminoglycoside-modifying enzymes. Micro-
biol Rev.; 57: 138-163.
Shimizu K, Kumada T, Hsieh WC et al. (1985) Comparison of
aminoglycoside resistance patterns in Japan, Formosa, and
Korea, Chile, and the United States. Antimicrob Agents Chem-
othe, 28: 282-288.