Genetic characterization of native
Bacillus thuringiensis
strains isolated from Tamil Nadu, India
D. Immanual Gilwax Prabhu
1
*, S. John Vennison
2
*, P. Thirumalai Vasan
3
and E. Kathiresan
2
1
Silkworm Breeding and Genetics, Central Tasar Research and Training Institute, Ranchi 835303, India
2
Department of Biotechnology, Anna University BIT-Campus, Tiruchirappalli 620024, India
3
Department of Biotechnology, Srimad Andavan College, Tiruchirappalli 620005, India
ABSTRACT
B. thuringiensis is a crystalliferous bacteria used as a biocontrol agent against lepidopteran, dipteran, and coleopteran
pests. Seventy eight Bacillus thuringiensis strains were isolated from 108 soil samples collected from Tamil Nadu, India.
Phylogenetic relationship of B. thuringiensis isolates were evaluated based on PCR ampli ed fragment polymorphisms
of agellin genes (PCR-AFPF). The isolated B. thuringiensis strains comprised of 51.3% of known biochemical types and
48.7% of undescribed B. thuringiensis types. PCR-AFPF UPGMA dendrogram generated using Jaccard coef cient val-
ues showed two phylogenetic groups, group A and B comprised of I-XIII and XIV-XV clusters respectively. The present
study concluded that B. thuringiensis isolates from Tamil Nadu have a high degree of genetic diversity and high rate of
genetic exchange.
KEY WORDS:
BACILLUS THURINGIENSIS
, PCR-AFPF, GENETIC DIVERSITY, UPGMA DENDROGRAM
587
Biotechnological
Communication
Biosci. Biotech. Res. Comm. 11(4): 587-594 (2018)
INTRODUCTION
Chemical insecticides that are currently used to control
insect pests are extremely toxic to non-target organisms
and many insects have developed resistance to differ-
ent chemical pesticides, resulting in ineffective insect
control programs. They are deleterious to the health of
humans and animals, lead to cancer and immune system
disorders. In addition, chemical insecticides are recalci-
trant, it accumulates in the environment and result in
soil and water pollution (Devine and Furlong, 2007). The
use of microbial insecticides is an alternative to chemi-
cal pesticides for insect control. Biological insecticides
are mainly based on entomatopathogenic bacteria,
Bacillus thuringiensis.
ARTICLE INFORMATION:
Corresponding Authors: immanual.csb@gmail.com
Received 13
th
Sep, 2018
Accepted after revision 13
th
Dec, 2018
BBRC Print ISSN: 0974-6455
Online ISSN: 2321-4007 CODEN: USA BBRCBA
Thomson Reuters ISI ESC / Clarivate Analytics USA
Mono of Clarivate Analytics and Crossref Indexed
Journal Mono of CR
NAAS Journal Score 2018: 4.31 SJIF 2017: 4.196
© A Society of Science and Nature Publication, Bhopal India
2018. All rights reserved.
Online Contents Available at:
http//www.bbrc.in/
DOI: 10.21786/bbrc/11.4/8

Immanual Gilwax Prabhu et al.
588 GENETIC CHARACTERIZATION OF NATIVE
BACILLUS THURINGIENSIS
STRAINS ISOLATED BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
B. thuringiensis is a member of a group of crystal-
liferous spore-forming gram-positive bacteria of the
family Bacillaceae (Schnepf et al., 1998). This bacte-
rium is able to produce proteinaceous parasporal crys-
tals that exhibit speci c insecticidal and nematicidal
activities (Khyami-Horani et al., 1996; Carneiro et al.,
1998; Al-Banna and Khyami-Horani, 2004). B. thur-
ingiensis is commonly used as an organic biopesticide
against lepidopteran, dipteran, and coleopteran insect
pests (Schnepf et al., 1998). B. thuringiensis was rst
isolated from diseased silkworm (Bombyx mori) larvae
(Ishiwata, 1901). Over the last ve decades, B. thuring-
iensis has been developed as a microbial agent against
lepidopteran pests (Carlton, 1990). In 1977, Goldberg
and Margalit (1977) discovered a novel B. thuringien-
sis strain that expressed speci c insecticidal properties
against Diptera and another B. thuringiensis strain that
targeted against Coleoptera was explored by Krieg et al.
(1983). The success of various B. thuringiensis strains in
controlling various insect pests has driven the establish-
ment of several screening programmes for more novel
B. thuringiensis strains. As a result, it is estimated that
today, more than 50,000 B. thuringiensis strains are kept
in various private and public bacterial collection centres
(Sanchis et al. 1996).
There are many methods have been proposed to clas-
sify B. thuringiensis into sub-species level. The main
classi cation of B. thuringiensis isolates was developed
on the basis of H- agellar antigens by de Barjac and
Bonnefoi (1962). B. thuringiensis strains are classi ed
into more than 82 serovars using H-antigen method.
However, there are two limitations with the H-classi-
cation for strains lacking parasporal inclusions and
auto-agglutinated strains. Some B. cereus strains have
antigens that cross-react with sera speci c for B. thur-
ingiensis H-serotypes (14) and such isolates may origi-
nate from older B. thuringiensis strains that have lost
plasmid encoded crystals (Lecadet et al., 1999). These
auto-agglutinated strains make up almost 3% of the B.
thuringiensis in the International Entomopathogenic
Bacillus Center (IEBC) collection (Burges et al., 1982) at
the Institute of Pasteur, Paris, France, but the H-classi -
cation is completely useless on these strains. In addition,
a few B. thuringiensis strains, called non-motile strains
such as B. thuringiensis var. wuhanensis, also escape
H-serotyping (de Barjac and Frachon, 1990).
With the development of molecular biology, the clas-
si cation and identi cation of bacteria has changed
from traditional phenotypic to genotypic methods in
recent decades. Pulsed eld gel electrophoresis (PFGE),
used for genotyping different bacterial strains of a spe-
ci c species as in the case of B. thuringiensis strains
(Gaviria and Priest, 2003). However, as PFGE requires
special equipment and chemicals, it is not easy to per-
form in many laboratories. Currently, molecular typing
methods including Arbitrary Primer PCR technology
(Brousseau et al., 1993), DNA reassociation measure-
ments (Nakamura, 1994), ribosomal RNA gene restric-
tion fragment length polymorphism (Priest et al., 1994;
Akhurst et al., 1997), ribosomal RNA gene intergenic
spacer sequences comparison (Bourque et al., 1995),
DNA-colony hybridization and random ampli ed pol-
ymorphic DNA (RAPD) analysis (Hansen et al., 1998),
have also been applied to a limited numbers of B. thur-
ingiensis strains. To date, molecular techniques such as
RAPD, RFLP, 16S rRNA probe, speci c DNA probe, and
ISR methods have not provided any great improvement
over the H-classi cation method. Hence, in the present
research, a faster, convenient and accurate method was
followed to classify all subspecies of B. thuringiensis
using PCR ampli ed fragment polymorphism of agel-
lin genes (PCR-AFPF).
MATERIALS AND METHODS
Soil collection and isolation of B. thuringiensis: Soil
samples were collected from 108 locations in Tamil
Nadu, India, that are very diverse in nature. The sam-
ples were collected from agricultural elds, high-altitude
mountains, forests, grasslands and sewage. Soil samples
were collected by scraping off surface material with a
sterile spatula and then obtaining a 10g sample from 1
or 2 cm below the surface. These samples were stored in
sterile plastic bags at ambient temperature. One gram of
soil was added to 10ml of Luria Broth, which was buff-
ered with 0.25M sodium acetate (Travers et al., 1987).
The mixture was shaken for 4h at 250 rpm in 30
o
C, after
incubation 1.5ml of sample mixture was taken and heat
shocked at 80
o
C for 3min. 100μl of the suspension was
plated on HiCrome™ Bacillus Agar supplemented with
10μg/ml of polymyxin B. Colonies formed after over-
night growth at 30ºC were selected based on colour
and colony morphology and the colony was transferred
onto TCHA medium supplemented with 0.3% glucose.
Cultures were allowed to grow and sporulate for 40h at
30ºC and the sporulated cultures were then checked for
the presence of crystals, which was the criterion used to
con rm the isolates as B. thuringiensis (Braun, 2000).
Biochemical identi cation
Fourteen biochemical tests such as acid production from
glucose, arabinose, xylose, mannitol, mannose, salicin
and sucrose; utilization of citrate and esculin; and pro-
duction of protease, amylase, phospholipase C or leci-
thinase, and hemolysin were performed as described by
Parry et al. (1983) to identify B. thuringiensis strains.
For this study, only the results of the following four (the
Immanual Gilwax Prabhu et al.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS GENETIC CHARACTERIZATION OF NATIVE
BACILLUS THURINGIENSIS
STRAINS ISOLATED 589
most relevant) biochemical tests are presented: esculin
utilization, acid formation from salicin and sucrose, and
lecithinase production (Martin and Travers, 1989). Based
on these four biochemical tests the B. thuringiensis iso-
lates classi ed into 16 biochemical types. The classi -
cation of B. thuringiensis strains in these groups was
corresponding to the distribution obtained by additional
tests.
PCR-AFPF
DNA C1000 thermal cycler (Bio-Rad) was used to carry
out PCR ampli cation. Cells of different B. thuringiensis
strains were inoculated on Luria-Bertani (LB) agar plates
and incubated at 30°C for 12 h. A loopful of cells was
suspended in 100 μl of nuclease free water in a 1.5-ml
Eppendorf tube. The cell suspension was frozen at -70
o
C
for 20 min and then boiled in water bath for 10 min.
The resultant lysate was centrifuged at 10,000 rpm for
10 min and ve microliters of the supernatant was used
as a source of DNA template. The AFPF primers used in
this study was previously described by Yu et al. (2002)
and are listed (Table 1). Primer was obtained from Euro-
ns MWG Operon, Germany. Each 50 μl of PCR mixture
contained 200 μM deoxynucleotide triphosphates, 2 μM
MgCl
2
, 12.5 pmol per primer, and 2 U of DreamTaq
TM
DNA polymerase (Fermentas). Ampli cation was per-
formed using a single denaturation of 3 min at 94°C
followed by a 35 cycle program, with each cycle con-
sisting of denaturation at 94°C for 30 s, annealing at
45°C for 30 s, and extension at 72°C for 2 min; the nal
extension step was 72°C for 10 min. The PCR ampli ed
products were detected by 1% agarose gel electrophore-
sis. The gels were scanned using AlphaImager gel docu-
mentation system.
Data analysis
Cluster analysis was used to examine genotypic rela-
tionships among the environmental B. thuringiensis
isolates and it was performed using the AlphaView soft-
ware, version 4.2 (Proteinsimple, CA). For data analy-
sis, the pro les were converted into binary matrix. The
computer cluster analysis was performed on the basis of
calculation of the Jaccard Coef cient using unweighted
pair group method with arithmetic mean (UPGMA)
(Kumar et al., 1993).
RESULTS AND DISCUSSION
Isolation of B. thuringiensis from soil: Totally 108 soil
samples were collected from different locations of 26
districts in Tamil Nadu, India. Soil samples were col-
lected from cultivated elds (rice, sugarcane, coffee, tea,
mango, papaya, cabbage, onion, tomato, coconut, and
potato), natural vegetation (pine forests, shola forest,
reserved forest, tropical evergreen forest, and grasslands)
and sewage soils. The elevations of the places from
which the samples collected were highly variable, rang-
ing from sea level to 7,200 m above sea level. Among
82 soil samples, twenty six samples were collected from
hilly regions (Ooty, Yercaud, Kolli hills, Palani, Kodaika-
nal, Sirumalai).
After acetate and polymyxin B selection, 97 of 108
soil sample yield colonies. Of these 97 samples, 70
(72%) contained at least one crystal protein forming B.
thuringiensis strains. From the 70 soil samples, 78 B.
thuringiensis strains were isolated and separated from
173 other spore forming organisms. Overall, this sug-
gests that Tamil Nadu soils were enriched with B. thur-
ingiensis.With the advent of a very selective procedure
for separating B. thuringiensis spores from the spores
of other soil microbes, 78 B. thuringiensis strains have
been isolated through acetate and polymyxin B selec-
tion. The distribution of B. thuringiensis in Tamil Nadu,
is summarized (Table 2). In Tamil Nadu, the soil samples
collected from districts such as, Vellore, Thanjavur and
Theni were extremely rich in B. thuringiensis. Hundred
percent of the soil sample contained B. thuringiensis.
The soil sample collected from Krishnagiri, Nilgiris,
Coimbatore, Trichy, Tirunelvelli and Kanyakumari had
75 – 80% of B. thuringiensis strains. On the other hand,
soil samples collected from Ariyalur, Nagapatinam,
Madurai, Virudhunagar and Tuticorin, only 25 – 33% of
the soil samples contained B. thuringiensis strains.
Biochemical typing: The B. thuringiensis strains iso-
lated from Tamil Nadu were identi ed and classi ed
based on Martin and Travers (1989). In order to dis-
criminate the isolated B. thuringiensis strains in to 16
biochemical types, four biochemical tests such as escu-
lin utilization, acid formation from salicin and sucrose
and lecithinase production were carried out and these
were the most variable among B. thuringiensis isolates.
B. thuringiensis isolates were given the biochemical
type number based on the Martin and Travers (1989)
and their occurrence in Tamil Nadu is also recorded
(Table 3). Eventhough some of these types were differed
by a single biochemical tests, these difference were more
important. In this study B. thuringiensis subsp. kurstaki
is widely available in the environment differed from
leastly available B. thuringiensis subsp. galleriae by a
single biochemical test, lecithinase production. Among
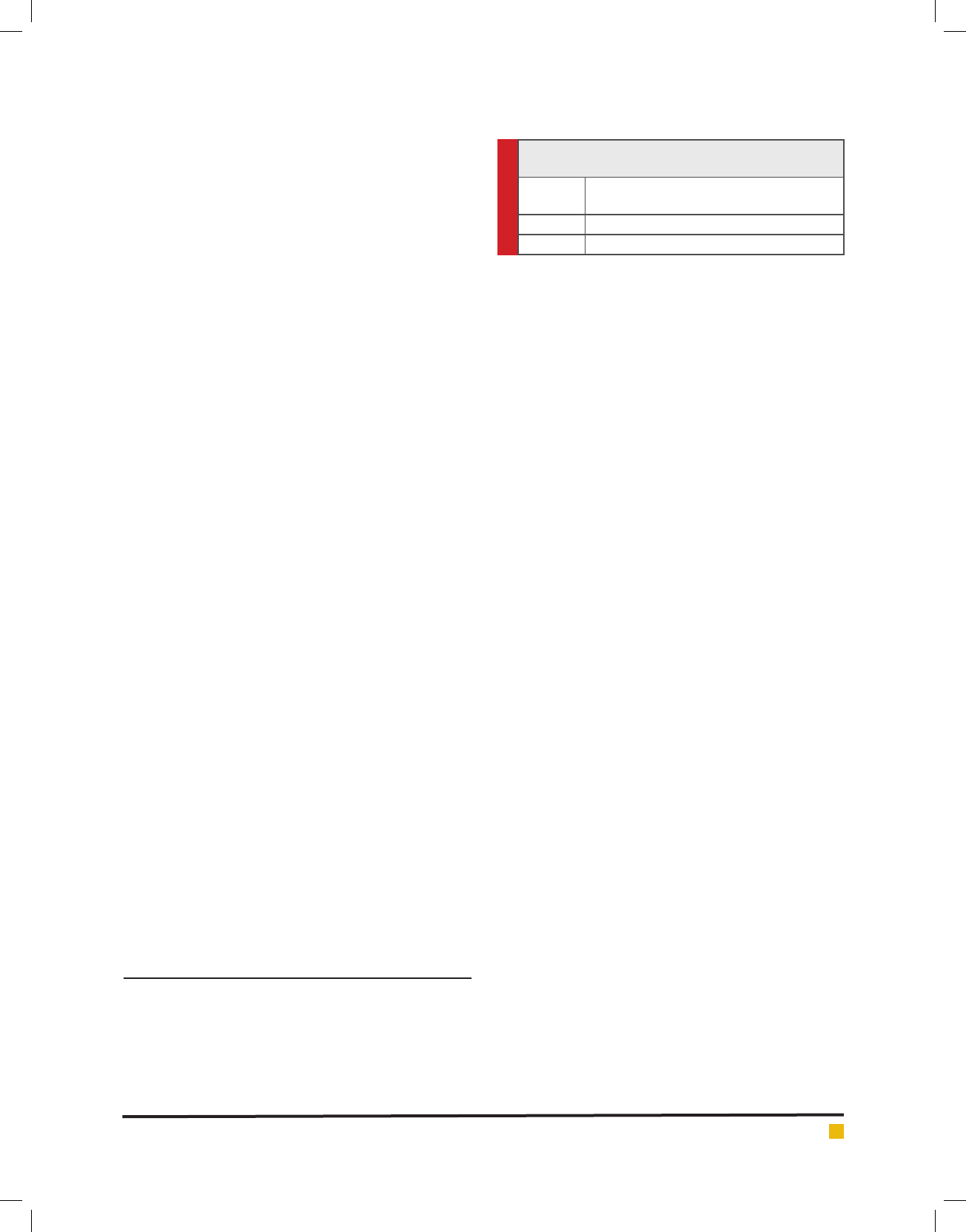
Table 1. Nucleotide sequences of primers used for
PCR-AFPF and multiplex PCR
Primer
Name Sequence
Fla5 F – GGCGTCGACATGAGAATTAATACAAACATT
Fla3 R – CGCCTGCAGTTATTGTAATAATTTAGAAGCC

Immanual Gilwax Prabhu et al.
590 GENETIC CHARACTERIZATION OF NATIVE
BACILLUS THURINGIENSIS
STRAINS ISOLATED BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
Table 2. Distribution of B. thuringiensis in soil
Districts
No. of samples
examined
% of samples with
B.
thuringiensis
isolate (no.
of samples)
B. thuringiensis
indexa
(no. of
B. thuringiensis
isolates)
Chennai 4 50.0 (2) 0.22 (2)
Kanchipuram 4 50.0 (2) 0.33 (2)
Vellore 4 100.0 (4) 0.37 (3)
Krishnagiri 4 75.0 (3) 0.28 (4)
Villupuram 4 50.0 (2) 0.18 (2)
Nilgiris 4 75.0 (3) 0.40 (4)
Erode 4 50.0 (2) 0.22 (2)
Salem 4 75.0 (3) 0.38 (5)
Ariyalur 4 25.0 (1) 0.16 (1)
Perambulur 4 50.0 (2) 0.25 (2)
Namakkal 4 75.0 (3) 0.46 (6)
Coimbatore 4 75.0 (3) 0.36 (4)
Tiruppur 4 50.0 (2) 0.28 (2)
Karur 4 75.0 (3) 0.25 (2)
Trichy 4 75.0 (3) 0.27 (3)
Nagapattinam 4 33.3 (1) 0.16 (1)
Tiruvarur 4 66.6 (2) 0.40 (2)
Thanjavur 4 100.0 (4) 0.30 (4)
Pudukottai 4 75.0 (3) 0.33 (2)
Dindigul 4 100.0 (4) 0.30 (4)
Theni 4 100.0 (4) 0.33 (4)
Madurai 4 75.0 (2) 0.44 (4)
Virudhunagar 4 50.0 (2) 0.12 (1)
Tuticorin 4 50.0 (2) 0.28 (2)
Tirunelveli 4 75.0 (3) 0.36 (4)
Kanyakumari 4 75.0 (3) 0.60 (3)
Ramanathapuram 4 50.0(2) 0.28 (2)
Total 108 64.8 (70) 0.31 (78)
a
B. thuringiensis index was calculated as a number of B. thuringiensis strains isolated divided by the number of
colonies of all bacteria examined.
78 B. thuringiensis isolates, a maximum of 12 (15.4%) B.
thuringiensis subsp. kurstaki strains were isolated from
different soil samples. B. thuringiensis subsp. israelensis
was not present in any of the samples. The biochemical
types 9, 10, 11, 13, 15 and 16 made up a cluster of strains
which accounted for 48.7% of all environmental iso-
lates. The biochemical type of each isolates is recorded
(Table 3). The results are similar to the study reported
by Martin and Travers (1989), but they have got more
isolates of six undescribed B. thuringiensis biochemical
types (52%) than the known biochemical types (48%).
The widely accepted and well established typing
method for B. thuringiensis strains is serotyping. But the
current serotyping system is not suitable for auto agglu-
tinated strains, non-motile strains and strains lacking
a parasporal inclusion body. From the point of bacte-
rial systematic classi cation, serotyping is a phenotypic
system which cannot reveal phylogenetic relationships
among the strains (Joung and Côté, 2001).
Analysis of AFPF-PCR ampli ed products: PCR was
performed using cell lysate as DNA template with Fla5
and Fla3 primers. AFPF-PCR yielded multiple distinct
DNA products of sizes ranging from approximately 100
to 2000bp of more than 300 fragments from 78 B. thur-
ingiensis strains (Fig. 1). All B. thuringiensis isolates dif-
fered from one another in the speci c ampli ed patterns
of the PCR products, which correspond to the presence of
agellin gene sequence. In these isolates the major num-
ber of bands was observed at the size of 100, 225 and
425bp. Most of the ampli ed products were observed

Immanual Gilwax Prabhu et al.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS GENETIC CHARACTERIZATION OF NATIVE
BACILLUS THURINGIENSIS
STRAINS ISOLATED 591
Table 3. Biochemical types of B. thuringiensis and its occurrence in Tamil Nadu, India
a
Biochemical Type
Biochemical test result
b
B. thuringiensis
isolates in Tamil
Nadu, India (%)Esculin Salicin Lecithinase Sucrose
1 (thuringiensis) + + + + 4 (5.1)
2 (kurstaki) + + + - 12 (15.4)
3 (indiana) + + - + 3 (3.8)
4 (galleriae) + + - - 1 (1.3)
5 (sotto) + - + + 3 (3.9)
6 (dendrolimus) + - + - 11 (14.1)
7 (morrisoni) + - - + 3 (3.8)
8 (darmstadiensis) + - - - 2 (2.6)
9 (Biochemical type 9) - + + + 4 (5.1)
10 (Biochemical type 10) - + + - 6 (7.7)
11 (Biochemical type 11) - + - + 1 (1.3)
12 (ostriniae) - + - - 1 (1.3)
13 (Biochemical type 13) - - + + 9 (11.5)
14 (israelensis) - - + - 0 (0)
15 (Biochemical type 15) - - - + 10 (12.8)
16 (Biochemical type 16) - - - - 8 (10.3)
Total 78
a
Martin and Travers, 1989
b
The + sign indicates a positive reaction; – sign indicates a negative reaction
FIGURE 1. PCR-AFPR ngerprint patterns of environmental isolates of Bacillus thur-
ingiensis. (A) Bacillus thuringiensis Tamil Nadu (BTTN) isolates BTTN01 – BTTN20; (B)
BTTN20 – BTTN40; (C) BTTN41 – BTTN60; (D) BTTN61 – BTTN78. Lane M, 100bp ladder.
Immanual Gilwax Prabhu et al.
592 GENETIC CHARACTERIZATION OF NATIVE
BACILLUS THURINGIENSIS
STRAINS ISOLATED BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
FIGURE 2. UPGMA dendrogram derived from similarity
coef cients calculated by the Jaccard method based on
the ampli ed bands obtained during PCR-AFPF analy-
sis. The dendrogram shows the relationships among
environmental isolates of B. thunringiensis.
between 100 to 1000bp and only few of the isolates
(BTTN4, 6, 16, 23, 44, 56 and 66) contained fragments
more than 1000bp. Only a distinct single product with a
size of about 100 or 425bp was obtained in the strains
such as, BTTN19, 20, 21, 29, 38, 40, 60 and 70. The max-
imum of 12 to 14 bands ranged from sizes 100 – 2000bp
were observed in BTTN4, 23, 56 and 66.
PCR-AFPF is a preliminary attempt of Yu et al. (2002)
to classify the subspecies of B. thuringiensis obtained
from the Institute of Pasteur, France and in the present
study this tool is used to discriminate between the B.
thuringiensis serovars isolated from the environment.
The chromosome of all B. thuringiensis strains contain
agellin gene and it was present even in their closely
related species B. cereus and B. anthracis. Its PCR ampli-
cation pattern analysis is not subjected to any limi-
tation associated with the B. thuringiensis serotyping
system. The phyolgenetic analysis of B. thuringiensis
using PCR-AFPF would be more accurate, than previ-
ously described molecular classi cation methods such
as RAPD, RFLP, 16s rRNA probe, speci c DNA probe,
and ISR (Yu et al., 2002). The AFPF primers recognises
differences in the prevalence and positions of anneal-
ing sites in the genome producing sets of fragments that
are considered to re ect the genomic composition of the
strain, therefore it gives a good opportunity to detect
biodiversity of a group of isolates.
Phylogenetic analysis using UPGMA method: The
genetic distance, Jaccard coef cient values were calcu-
lated from ampli ed bands and the values were ranged
from 0.07 to 0.035 (Fig. 2). An unweighed pair group
method with average (UPGMA) dendrogram was con-
structed using the Jaccard coef cient. In UPGMA den-
drogram, the B. thuringiensis isolates were divided into
2 groups, group A and B. Group A was further classi-
ed into 13 clusters (I - XIII), which comprised of 68 B.
thuringiensis isolates and remaining 10 strains, BTTN71,
33, 22, 39, 67, 32, 29, 18, 26 and 73 were categorized in
cluster XIV and XV and these two clusters were grouped
under group B. Among I to XV clusters, cluster I, X, XII
and XIV contains the mixture of different B. thuring-
iensis biochemical types and rest of the 11 clusters con-
tains any one of the biochemical types. The maximum
of 20.5% of B. thuringiensis isolates were classi ed
under cluster X and it comprised 6.25% of biochemi-
cal type 9, 25% of B. thuringiensis subsp. kurstaki and
68.75% of B. thuringiensis subsp. dendrolimus. At the
least of cluster XIII contained only one biochemical type
9 strain and this type showed more degree of variations
in their band pattern among their biochemical type and
these four biochemical type 9 strains were characterized
under 3 clusters (X, XI and XIII). This con rmation of
variation or intermixing of biochemical type 9 within
the clusters suggests that the speci c phenotypes were
acquired after the ancestors to each of the clusters were
formed. The study supports the idea that horizontal gene
transfer of plasmid is an important factor in de ning the
phenotypes of biochemical type 9 isolates evolved along
perceptible large evolutionary distances to give rise to
different clusters.
The study of Katara et al. (2012) demonstrated that
molecular typing and diversity analysis of B. thuringien-
sis has enormous importance for discrimination of strains
isolated from different sources. They distinguished 113
native B. thuringiensis strains isolated from various

Immanual Gilwax Prabhu et al.
BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS GENETIC CHARACTERIZATION OF NATIVE
BACILLUS THURINGIENSIS
STRAINS ISOLATED 593
locations in India using REP-PCR and ERIC-PCR. They
explored that the B. thuringiensis isolates collected from
diverse habitats in India had a high degree of genetic
diversity. Similar to them, the B. thuringiensis strains
which were isolated from Tamil Nadu showed diverse
range of patterns and high level of genetic diversity.
CONCLUSION
Through the present study, it is suggested that the asso-
ciation between B. thuringiensis and insects is not obli-
gative. B. thuringiensis was omnipresent when com-
pared to other bacterial strains and based on the nutrient
requirements the growth turnover is possible. There is
no need that the population of B. thuringiensis has to be
more in the soil samples with high levels of insect activ-
ity because several soil samples had been collected from
the mosquito breeding regions but no B. thuringiensis
isolates had shown any degree of mosquito larvicidal
activity. Finally, the genetic heterogeneity were analysed
in the B. thuringiensis strains isolated from Tamil Nadu
using AFPF-PCR. This technique could be used for the
separation of novel B. thuringiensis isolates from the
environment.
REFERENCES
Akhurst, R.J., Lyness, E.W., Zhang, Q.Y., Cooper, D.J., Pinnock,
D.E. (1997) A 16S rRNA oligonucleotide probe for identi ca-
tion of Bacillus thuringiensis isolates from sheep eece. J.
Invertebr. Pathol. 69, 24–31.
Al-Banna, L., Khyami-Horani, H. (2004) Nematicidal activity
of two Jordanian strains of Bacillus thuringiensis on root-knot
nematodes. Nematol. Mediterr. 32, 41–45.
Bourque, S.N., Valero, J.R., Lavoie, M.C., LeÂvesque, R.C. (1995)
Comparative analysis of the 16S to 23S ribosomal intergenic
spacer sequences of Bacillus thuringiensis strains and subspe-
cies and of closely related species. Appl. Environ. Microbiol.
61, 1623-1626.
Braun, S. (2000) Production of Bacillus thuringiensis insec-
ticides for experimental uses. In: Navon, A., Ascher, K.R.S.
(Eds.), Bioassays of Entomopathogenic Microbes and Nema-
todes, CABI, London, UK, pp. 49-72.
Brousseau, R., Saint-Onge, A., PreÂfontaine, G., Masson, L.
And Cabana, J. (1993) Arbitrary primer polymerase chain reac-
tion, a powerful method to identify Bacillus thuringiensis sero-
vars and strains. Appl. Environ. Microbiol. 59, 114-119.
Burges, H.D., Aizawai, A., Dulmage, H.T., de Barjac, H. (1982)
Numbering of the H-serotypes of Bacillus thuringiensis. J.
Invertebr. Pathol. 40, 419.
Carlton, B. (1990) Alternatives for suppressing agricultural
pests and diseases. In: Baker, R.R., Dunn, P.E., Liss, A.R. (Eds.),
New Directions in Biological Control. New York: American
Elsevier Publishing Co., pp. 419-434.
Carneiro, R.M.D.G., de Souza, I.S., Belarmino, L.C. (1998)
Nematicidal activity of Bacillus spp. strains on juveniles of
Meloidogyne javanica. Nematol. Brasileira. 22, 12–21.
de Barjac, H., Bonnefoi, A. (1962) Essai de classi cation bio-
chimique et serologique de 24 souches de Bacillus du type B.
thuringiensis. Entomophaga 7, 5–31
de Barjac, H., Frachon, E. (1990) Classi cation of Bacillus
thuringiensis strains. Entomophaga 35, 233–240
Devine, G.J., Furlong, M.J. (2007) Insecticide use: contexts and
ecological consequences. Agric. Human Values 24, 281–306.
Gaviria, R.A.M., Priest, F.G. (2003) Pulsed eld gel electropho-
resis of chromosomal DNA reveals a clonal population struc-
ture to Bacillus thuringiensis that relates in general to crystal
protein gene content. FEMS Microbiol. Lett. 223, 61– 66.
Goldberg, L.Y., Margalit, J. (1977) A bacterial spore demon-
strating rapid larvicidal activity against Anopheles sergentii,
Uranotaenia unguiculata, Culex univittatus, Aedes aegypti and
Culex pipiens. Mosq. News 37, 355-358.
Hansen, B., Damgaard, P.H., Eilenberg, J., Pedersen, J.C. (1998)
Molecular and phenotypic characterization of Bacillus thur-
ingiensis isolated from leaves and insects. J. Invertebr. Pathol.
71, 106-114.
Ishiwata, S. (1901) On a kind of severe acherue (sotto disease).
Dainihan Sanshi Kaiho 114, 1-5.
Joung, K.B., Côté, J.C. (2001) Phylogenetic analysis of Bacil-
lus thuringiensis serovars based on 16S rRNA gene restriction
fragment length polymorphisms. J. Appl. Microbiol. 90, 115-22.
Katara, J., Deshmukh, R., Singh, N.K., Kaur, S. (2012) Molecular
typing of native Bacillus thuringiensis isolates from diverse
habitats in India using REP-PCR and ERIC-PCR analysis. J.
Gen. Appl. Microbiol. 58, 83-94.
Khyami-Horani, H., Katbeh-Bader, A., Mohsen, Z.H. (1996)
Mosquito larvicidal toxicity of endospore-forming bacilli iso-
lated in Jordan. Dirasat. Med. Biol. Sci. 23,140–144
Krieg, A., Huger, A.M., Langenbruch, G.A., Schnetter, W. (1983)
Bacillus thuringiensis
var. tenebrionis, a new pathotype effec-
tive against larvae of Coleoptera. Zeitschrift für Angewandte
Entomologie 96, 500 - 508.
Kumar, S., Tamura, K., Nei, M. (1993) MEGA: Molecular Evolu-
tionary Genetics Analysis, version 1.0 (user manual), The Penn-
sylvania State University, University Park, PA 16802, USA.
Lecadet, M.M., Frachon, E., Dumanoir, V.C., Ripouteau, H.,
Hamon, S., Laurent, P., Thiery, I. (1999) Updating the H-anti-
gen classi cation of Bacillus thuringiensis. J. Appl. Microbiol.
86, 660–672.
Martin, P.A.W., Travers, R.S. (1989) Worldwide Abundance and
Distribution of Bacillus thuringiensis Isolates. Appl. Environ.
Microbiol., 55, 2437-2442.
Nakamura, L.K. (1994) DNA relatedness among Bacillus thur-
ingiensis serovars. Int. J. Syst. Bacteriol. 44, 125-129.
Parry, J.M., Turnball, P.C.B., Gibson, J.R. (1983) A Colour Atlas
of Bacillus Species. Wolfe Medical Publications, Ltd., London.
United Kingdom.

Immanual Gilwax Prabhu et al.
594 GENETIC CHARACTERIZATION OF NATIVE
BACILLUS THURINGIENSIS
STRAINS ISOLATED BIOSCIENCE BIOTECHNOLOGY RESEARCH COMMUNICATIONS
Priest, F.G., Kaji, D.A., Rosato, Y.B., Canhos, V.P. (1994) Char-
acterization of Bacillus thuringiensis and related bacteria by
ribosomal RNA gene restriction fragment length polymor-
phisms. Microbiology 140, 1015-1022.
Sanchis, V., Chaufaux, J., Lereclus, D. (1996) Amélioration
biotechnologique de Bacillus thuringiensis: les enjeux et les
risques. Annales de l’Institut Pasteur/Actualités 7, 271-284.
Schnepf, E., Crickmore, N., Van Rie, J., Lereclus, D., Baum, J.,
Feitelson, J., Zeigler, D.R., Dean, D.H. (1998) Bacillus thuring-
iensis and its pesticidal crystal proteins. Microbiol. Mol. Biol.
Rev. 62, 775–806.
Travers, R.S., Martin, P.A.W., Reichelderfer, C.F. (1987) Selec-
tive process for ef cient isolation of soil Bacillus sp. Appl.
Environ. Microbiol. 53, 1263-1266.
Yu, J., Tan, L., Liu, Y., Pang, Y. (2002) Phylogenetic analy-
sis of Bacillus thuringiensis based on PCR ampli ed fragment
polymorphisms of agellin genes. Curr. Microbiol. 45, 139-
43.